Agilent data
This analysis was performed using R (ver. 3.1.0).Download raw data
The raw data files for this lab are in the rawdata repository, available here:
https://github.com/genomicsclass/rawdata.
Click Download ZIP in order to download all the files, unzip this file which should result in a rawdata-master folder. Rename this folder to rawdata.
Read Agilent data
Agilent data is a two color arrays. For two-color arrays it’s slightly more complicated, because you have a pairing of files (red and green channels). Different scanners spit out different formats for this. So the target information will be a little bit different. limma package is used to read the files. The readTargets function tells you what’s in the red and the green channels.
The function read.maimages(microarray images) is used to read the data. You have to tell it what software produced the images. There’s different software for that, although these days, there’s a default that works pretty broadly. So all we’re doing now is telling it where the files are. We tell it what imaging software was used to produce these files (“genepix”). The function read.maimages, stores red and green separately.
library(limma)
library(rafalib)#installed from github
#Define a base directory
basedir <- "~/hubiC/Documents/R/doc/english/genomics/rawdata/agilent"
setwd(basedir)
#Read sample information table : it tells you what's in the red channel and the green channel
targets <- readTargets("TargetBeta7.txt")
#Read microarray files : Red and green are stored separately
RG <- read.maimages(targets$FileName, source="genepix")## Read 6Hs.195.1.gpr
## Read 6Hs.168.gpr
## Read 6Hs.166.gpr
## Read 6Hs.187.1.gpr
## Read 6Hs.194.gpr
## Read 6Hs.243.1.gprNormalization
Data are normalized using MA.RG function. It stores the information as the log ratio– that’s the M– and the average of the logs, which is the A.
#Normalization
MA <- MA.RG(RG,bc.method="none")MA plot
Just to give you a quick idea of what you can do with this. You can do an MA plot. You just plot the A versus the M.
#Normalization
MA <- MA.RG(RG,bc.method="none")
#MA plot
plot(MA$A[,1],MA$M[,1])#MA plot for the first sample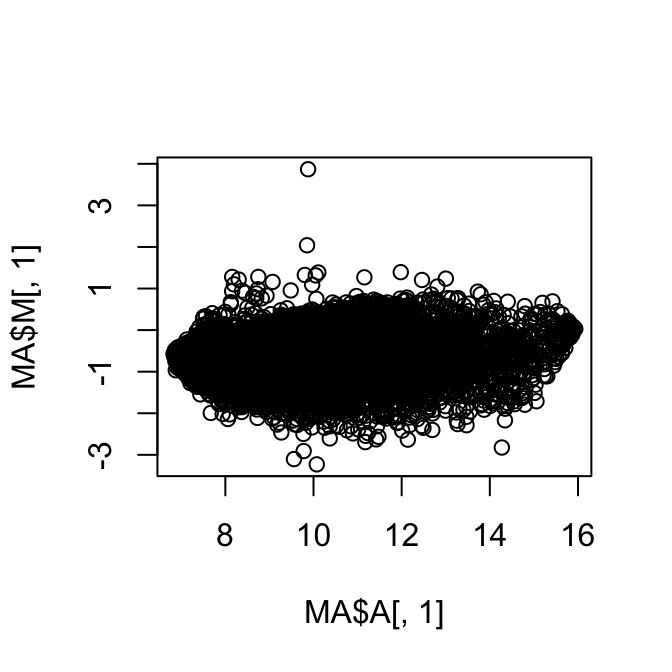
Array image
Another nice feature is to make images pretty quickly.
#Array image
imageplot(MA$M[,2], RG$printer, zlim=c(-3,3))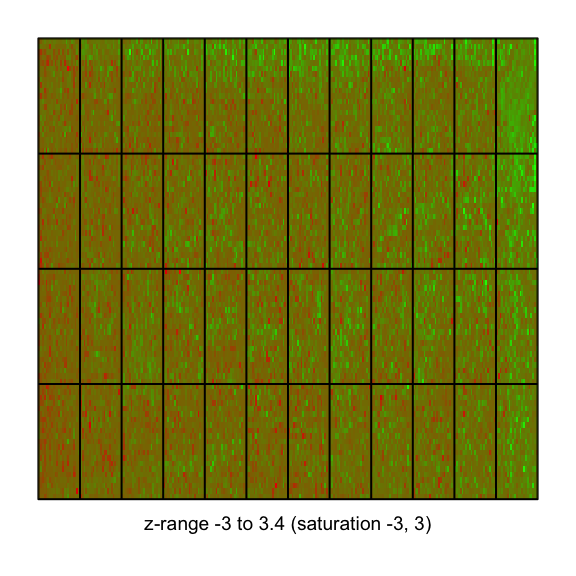
This is a nice example where we see an image where there seems to be a little bit of a problem. If you look at the edge, you see that it's quite green, compared to the middle, which is redder. Then you have too much red on the left. So we're detecting that there's somewhat of a problem in this file.Licence
References
Show me some love with the like buttons below... Thank you and please don't forget to share and comment below!!
Montrez-moi un peu d'amour avec les like ci-dessous ... Merci et n'oubliez pas, s'il vous plaît, de partager et de commenter ci-dessous!
Recommended for You!
Recommended for you
This section contains best data science and self-development resources to help you on your path.
Coursera - Online Courses and Specialization
Data science
- Course: Machine Learning: Master the Fundamentals by Standford
- Specialization: Data Science by Johns Hopkins University
- Specialization: Python for Everybody by University of Michigan
- Courses: Build Skills for a Top Job in any Industry by Coursera
- Specialization: Master Machine Learning Fundamentals by University of Washington
- Specialization: Statistics with R by Duke University
- Specialization: Software Development in R by Johns Hopkins University
- Specialization: Genomic Data Science by Johns Hopkins University
Popular Courses Launched in 2020
- Google IT Automation with Python by Google
- AI for Medicine by deeplearning.ai
- Epidemiology in Public Health Practice by Johns Hopkins University
- AWS Fundamentals by Amazon Web Services
Trending Courses
- The Science of Well-Being by Yale University
- Google IT Support Professional by Google
- Python for Everybody by University of Michigan
- IBM Data Science Professional Certificate by IBM
- Business Foundations by University of Pennsylvania
- Introduction to Psychology by Yale University
- Excel Skills for Business by Macquarie University
- Psychological First Aid by Johns Hopkins University
- Graphic Design by Cal Arts
Books - Data Science
Our Books
- Practical Guide to Cluster Analysis in R by A. Kassambara (Datanovia)
- Practical Guide To Principal Component Methods in R by A. Kassambara (Datanovia)
- Machine Learning Essentials: Practical Guide in R by A. Kassambara (Datanovia)
- R Graphics Essentials for Great Data Visualization by A. Kassambara (Datanovia)
- GGPlot2 Essentials for Great Data Visualization in R by A. Kassambara (Datanovia)
- Network Analysis and Visualization in R by A. Kassambara (Datanovia)
- Practical Statistics in R for Comparing Groups: Numerical Variables by A. Kassambara (Datanovia)
- Inter-Rater Reliability Essentials: Practical Guide in R by A. Kassambara (Datanovia)
Others
- R for Data Science: Import, Tidy, Transform, Visualize, and Model Data by Hadley Wickham & Garrett Grolemund
- Hands-On Machine Learning with Scikit-Learn, Keras, and TensorFlow: Concepts, Tools, and Techniques to Build Intelligent Systems by Aurelien Géron
- Practical Statistics for Data Scientists: 50 Essential Concepts by Peter Bruce & Andrew Bruce
- Hands-On Programming with R: Write Your Own Functions And Simulations by Garrett Grolemund & Hadley Wickham
- An Introduction to Statistical Learning: with Applications in R by Gareth James et al.
- Deep Learning with R by François Chollet & J.J. Allaire
- Deep Learning with Python by François Chollet







