IRanges
This analysis was performed using R (ver. 3.1.0).IRanges
IRanges is a library for representing ranges of integers, which is useful in genomics, because we have base pair ranges that we'd like to manipulate.There's a very detailed vignette on IRanges. IRanges have a start, an end and a width.
library(IRanges)
#IR with 6 base-pairs long : start and end are indicated
ir <- IRanges(5,10)
ir## IRanges of length 1
## start end width
## [1] 5 10 6start(ir) #Return the start value## [1] 5end(ir) #Return the end value## [1] 10width(ir) #return the width## [1] 6# ?IRangesIf you specify that the start is 5, and the width should be 6, you'll get the identical IRange.
#IR with 6 base-pairs long : start and width are indicated
ir <- IRanges(5,width=6)
ir## IRanges of length 1
## start end width
## [1] 5 10 6You can also specify more than one range at a time. If you give three starts and three ends, you get a IRanges object of length 3. If you, again, ask for the starts, you'll get the three different starts, 3, 5, and 17.
# Miultiple ranges
ir <- IRanges(start=c(3,5,17), end=c(10,8,20))
ir## IRanges of length 3
## start end width
## [1] 3 10 8
## [2] 5 8 4
## [3] 17 20 4length(ir)## [1] 3start(ir)## [1] 3 5 17There are a number of intra range methods for IRanges. And intra-range means that the operation will occur for each range that you have. An example of this is to shift the IRange to the left by two. Before we had an IRange that started at 5 and ended at 10. Applying the shift operation produces an IRange which starts at 3 and ends at 8.
# ?"intra-range-methods"
ir <- IRanges(5,10)
shift(ir, -2)## IRanges of length 1
## start end width
## [1] 3 8 6Remember, all of these commands can work on more than one range at once.Here we show the effects of the different methods using a single range:
#Shift the IRange to the left by 2 base-pairs
shift(ir,-2)## IRanges of length 1
## start end width
## [1] 3 8 6#narrow : Relative to the start, you should start this range at the second base pair.
narrow(ir, start=2)## IRanges of length 1
## start end width
## [1] 6 10 5#narrow: Relative to 5, you should end on the fifth base pair, which means it should end at 9.
narrow(ir, end=5)## IRanges of length 1
## start end width
## [1] 5 9 5#flank allows you to get flanking sequence
#here three base pairs from the start.
flank(ir, width=3, start=TRUE, both=FALSE)## IRanges of length 1
## start end width
## [1] 2 4 3#flank: three base pairs from the end by specifying start equals false
flank(ir, width=3, start=FALSE, both=FALSE)## IRanges of length 1
## start end width
## [1] 11 13 3#bidirectional flanking sequence from the start by specifying both equals true.
flank(ir, width=3, start=TRUE, both=TRUE)## IRanges of length 1
## start end width
## [1] 2 7 6ir * 2## IRanges of length 1
## start end width
## [1] 6 8 3ir + 2## IRanges of length 1
## start end width
## [1] 3 12 10ir - 2## IRanges of length 1
## start end width
## [1] 7 8 2The exact same functions are shown graphically with the code below:
# set up a plotting window so we can look at range operations
plotir <- function(ir,i) { arrows(start(ir)-.5,i,end(ir)+.5,i,code=3,angle=90,lwd=3) }
plot(0,0,xlim=c(0,15),ylim=c(0,11),type="n",xlab="",ylab="",xaxt="n")
axis(1,0:15)
abline(v=0:30 + .5,col=rgb(0,0,0,.5))
# plot the original IRange
plotir(ir,1)
# draw a red shadow for the original IRange
polygon(c(start(ir)-.5,start(ir)-.5,end(ir)+.5,end(ir)+.5),c(-1,12,12,-1),col=rgb(1,0,0,.2),border=NA)
plotir(shift(ir,-2), 2)
plotir(narrow(ir, start=2), 3)
plotir(narrow(ir, end=5), 4)
plotir(flank(ir, width=3, start=TRUE, both=FALSE), 5)
plotir(flank(ir, width=3, start=FALSE, both=FALSE), 6)
plotir(flank(ir, width=3, start=TRUE, both=TRUE), 7)
plotir(ir * 2, 8)
plotir(ir + 2, 9)
plotir(ir - 2, 10)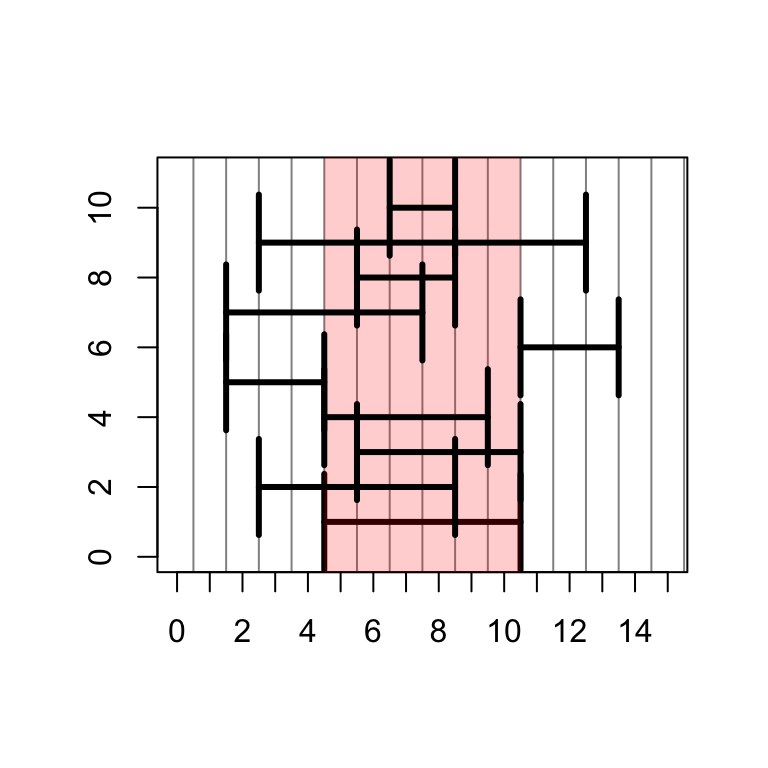
The inter-range methods are those functions which depend on the other ranges in the object. Let's create an IRanges object with three ranges, which starts at 3, 5, 17, ends at 10, 8, and 20.
# ?"inter-range-methods"
ir <- IRanges(start=c(3,5,17), end=c(10,8,20))
#range: returns the beginning of the IRanges to the end, including gaps in between.
range(ir)## IRanges of length 1
## start end width
## [1] 3 20 18#reduce, returns those base pairs which are covered by the original ranges.
#So we do not get the gap, so the end at 10 and the beginning at 17.
reduce(ir)## IRanges of length 2
## start end width
## [1] 3 10 8
## [2] 17 20 4#gaps: returns a gap from 10 to 17.
gaps(ir)## IRanges of length 1
## start end width
## [1] 11 16 6#disjoint: gives a set of ranges which has the same coverage as the original IRanges object,
#but they're not overlapping in any way, and they also contain the union of all the endpoints of the original range.
disjoin(ir)## IRanges of length 4
## start end width
## [1] 3 4 2
## [2] 5 8 4
## [3] 9 10 2
## [4] 17 20 4Licence
References
Show me some love with the like buttons below... Thank you and please don't forget to share and comment below!!
Montrez-moi un peu d'amour avec les like ci-dessous ... Merci et n'oubliez pas, s'il vous plaît, de partager et de commenter ci-dessous!
Recommended for You!
Recommended for you
This section contains the best data science and self-development resources to help you on your path.
Books - Data Science
Our Books
- Practical Guide to Cluster Analysis in R by A. Kassambara (Datanovia)
- Practical Guide To Principal Component Methods in R by A. Kassambara (Datanovia)
- Machine Learning Essentials: Practical Guide in R by A. Kassambara (Datanovia)
- R Graphics Essentials for Great Data Visualization by A. Kassambara (Datanovia)
- GGPlot2 Essentials for Great Data Visualization in R by A. Kassambara (Datanovia)
- Network Analysis and Visualization in R by A. Kassambara (Datanovia)
- Practical Statistics in R for Comparing Groups: Numerical Variables by A. Kassambara (Datanovia)
- Inter-Rater Reliability Essentials: Practical Guide in R by A. Kassambara (Datanovia)
Others
- R for Data Science: Import, Tidy, Transform, Visualize, and Model Data by Hadley Wickham & Garrett Grolemund
- Hands-On Machine Learning with Scikit-Learn, Keras, and TensorFlow: Concepts, Tools, and Techniques to Build Intelligent Systems by Aurelien Géron
- Practical Statistics for Data Scientists: 50 Essential Concepts by Peter Bruce & Andrew Bruce
- Hands-On Programming with R: Write Your Own Functions And Simulations by Garrett Grolemund & Hadley Wickham
- An Introduction to Statistical Learning: with Applications in R by Gareth James et al.
- Deep Learning with R by François Chollet & J.J. Allaire
- Deep Learning with Python by François Chollet
Click to follow us on Facebook :
Comment this article by clicking on "Discussion" button (top-right position of this page)







