Gviz - Visualize genomic data
This analysis was performed using R (ver. 3.1.0).Introduction to Gviz package
The Gviz package aims to provide a structured visualization framework to plot any type of data along genomic coordinates. It also allows to integrate publicly available genomic annotation data from sources like UCSC or ENSEMBL.
Individual types of genomic features or data are represented by separate tracks, like most of genome browsers.
By default, Gviz checks all supplied chromosome names for validity in the sense of the UCSC definition (chromosomes have to start with the chr string). You may decide to turn this feature off by calling options(ucscChromosomeNames=FALSE)
In the following examples, we will make use of the UCSC genome and chromosome 7 (chr7) on mouse mm9 genome.
Plot annotation track
Please note that the AnnotationTrack constructor can accommodate many different types of inputs. For instance, the start and end coordinates of the annotation features could be passed in as individual arguments start and end, as a data.frame or even as an IRanges or GRangesList object.
library(Gviz)
library(GenomicRanges)
#Load data : class = GRanges
data(cpgIslands)
cpgIslands## GRanges with 10 ranges and 0 metadata columns:
## seqnames ranges strand
##
## [1] chr7 [26549019, 26550183] *
## [2] chr7 [26564119, 26564500] *
## [3] chr7 [26585667, 26586158] *
## [4] chr7 [26591772, 26593309] *
## [5] chr7 [26594192, 26594570] *
## [6] chr7 [26623835, 26624150] *
## [7] chr7 [26659284, 26660352] *
## [8] chr7 [26721294, 26721717] *
## [9] chr7 [26821518, 26823297] *
## [10] chr7 [26991322, 26991841] *
## ---
## seqlengths:
## chr7
## NA #Annotation track, title ="CpG"
atrack <- AnnotationTrack(cpgIslands, name = "CpG")
plotTracks(atrack)
Add genome axis track
This step is to indicate the genomic coordinates we are currently looking at.
## genomic coordinates
gtrack <- GenomeAxisTrack()
plotTracks(list(gtrack, atrack))
Add chromosome ideogram
To add chromosome ideogram, we have to indicate a valid UCSC genome (e.g : “hg19”) chromosome name (e.g : “chr7”).
Internet connection is required as the function fetches data from UCSC and it can take quite long time.#genome : "hg19"
gen<-genome(cpgIslands)
#Chromosme name : "chr7"
chr <- as.character(unique(seqnames(cpgIslands)))
#Ideogram track
itrack <- IdeogramTrack(genome = gen, chromosome = chr)
plotTracks(list(itrack, gtrack, atrack))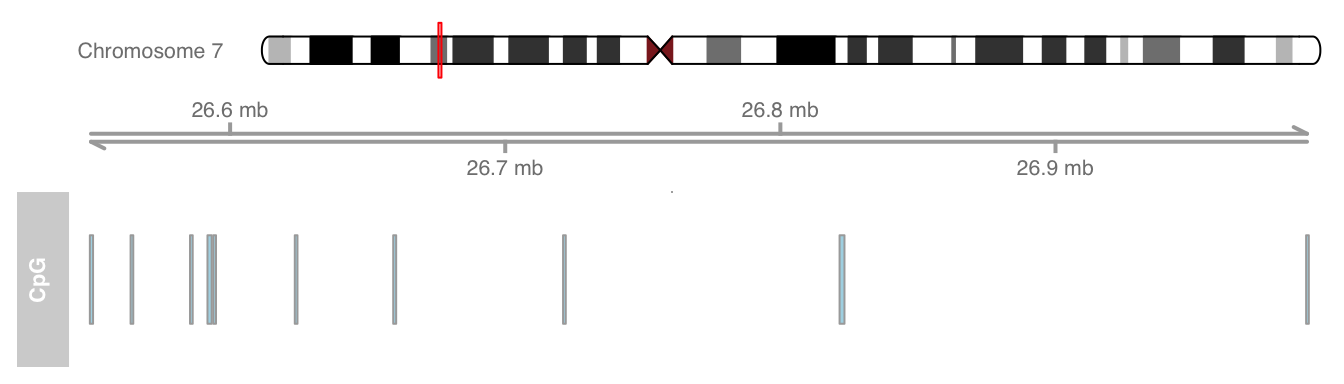
Ideogram tracks are the one exception in all of Gviz 's track objects in the sense that they are not really displayed on the same coordinate system like all the other tracks. Instead, the current genomic location is indicated on the chromosome by a red box (or, as in this case, a red line if the width is too small to fit a box).Add gene model
We can utilize gene model information from an existing local source. Alternatively, we could download such data from one of the available online resources like UCSC or ENSEBML, and there are constructor functions to handle these tasks.
For this example we are going to load gene model data from a stored data.frame.
#Load data
data(geneModels)
head(geneModels)## chromosome start end width strand feature gene exon transcript symbol
## 1 chr7 26591441 26591829 389 + lincRNA ENSG00000233760 ENSE00001693369 ENST00000420912 AC004947.2
## 2 chr7 26591458 26591829 372 + lincRNA ENSG00000233760 ENSE00001596777 ENST00000457000 AC004947.2
## 3 chr7 26591515 26591829 315 + lincRNA ENSG00000233760 ENSE00001601658 ENST00000430426 AC004947.2
## 4 chr7 26594428 26594538 111 + lincRNA ENSG00000233760 ENSE00001792454 ENST00000457000 AC004947.2
## 5 chr7 26594428 26596819 2392 + lincRNA ENSG00000233760 ENSE00001618328 ENST00000420912 AC004947.2
## 6 chr7 26594641 26594733 93 + lincRNA ENSG00000233760 ENSE00001716169 ENST00000457000 AC004947.2#Plot
grtrack <- GeneRegionTrack(geneModels, genome = gen,
chromosome = chr, name = "Gene Model")
plotTracks(list(itrack, gtrack, atrack, grtrack))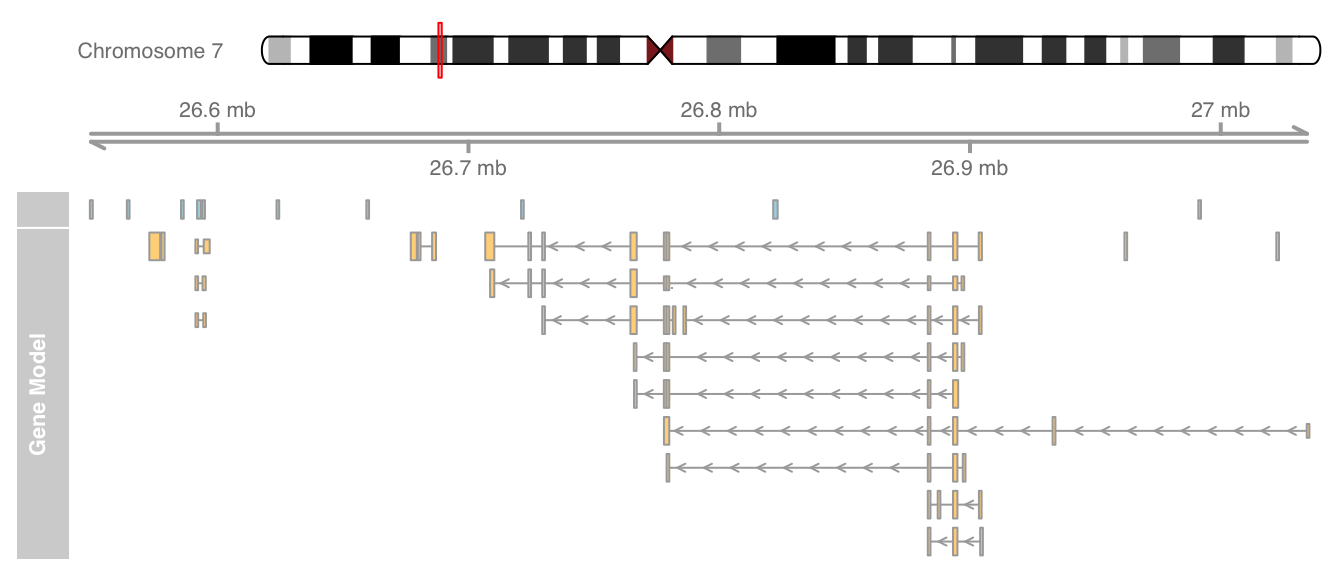
Zoom the plot
We often want to zoom in or out on a particular plotting region to see more details or to get a broader overview.
#Use from and to arguments to zoom
plotTracks(list(itrack, gtrack, atrack, grtrack),
from = 26700000, to = 26750000)
# Use extend.left and extend.right to zoom
#those arguments are relative to the currently displayed ranges,
#and can be used to quickly extend the view on one or both ends of the plot.
plotTracks(list(itrack, gtrack, atrack, grtrack),
extend.left = 0.5, extend.right = 1000000)
# to drop the bounding borders of the exons and
# to have a nice plot
plotTracks(list(itrack, gtrack, atrack, grtrack),
extend.left = 0.5, extend.right = 1000000, col = NULL)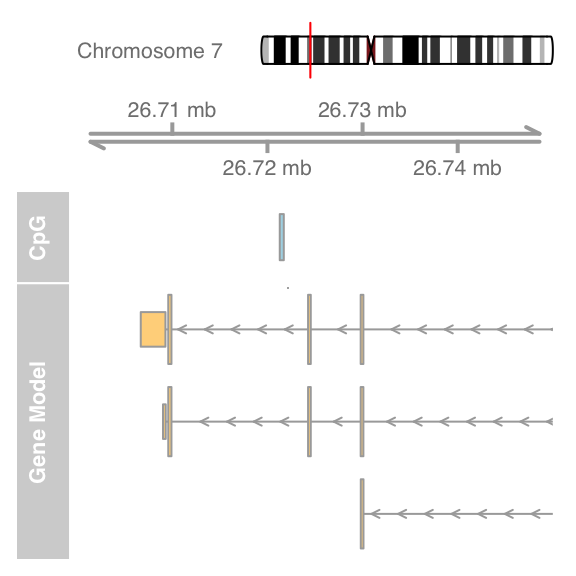
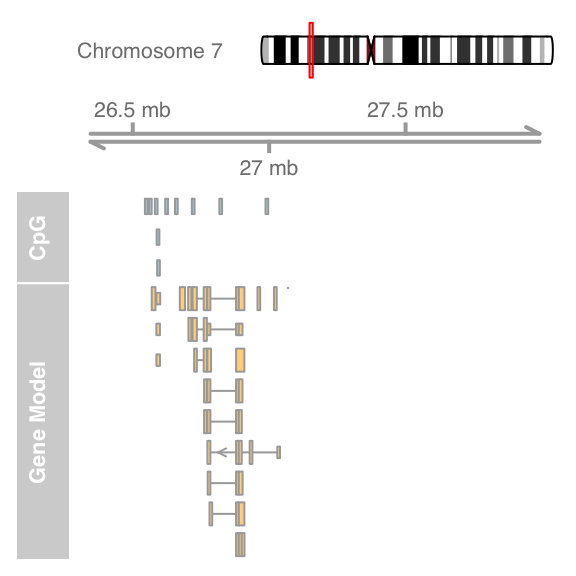
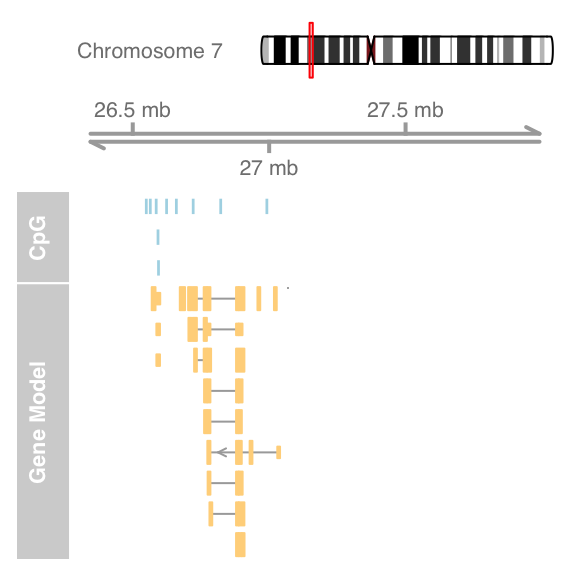
A value of 0.5 will cause zooming in to half the currently displayed range.Add sequence track and zoom to view sequence
The necessary sequence information is drawn from one of the BSgenome packages.
library(BSgenome.Hsapiens.UCSC.hg19)
strack <- SequenceTrack(Hsapiens, chromosome = chr)
plotTracks(list(itrack, gtrack, atrack, grtrack,
strack), from = 26591822, to = 26591852, cex = 0.8)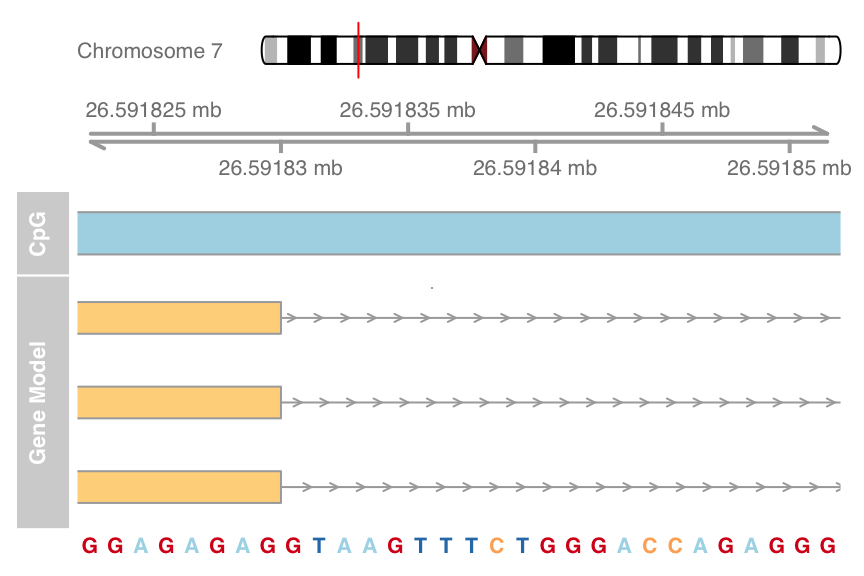
Add data track
DataTrack object are essentially run-length encoded (Rle) numeric vectors or matrices, and we can use them to add all sorts of numeric data to our genomic coordinate plots. Different visualization options for these tracks are available including dot plots, histograms and box-and-whisker plots.
#For demonstration purposes we can create a simple DataTrack object from
#randomly sampled data.
set.seed(255)
lim <- c(26700000, 26750000)
coords <- sort(c(lim[1], sample(seq(from = lim[1],
to = lim[2]), 99), lim[2]))
dat <- runif(100, min = -10, max = 10)
head(dat)## [1] 3.5382 -9.3984 -2.8372 -1.9871 0.2722 8.2508##data track
dtrack <- DataTrack(data = dat, start = coords[-length(coords)],
end = coords[-1], chromosome = chr, genome = gen,
name = "Uniform")
##Plot data track
plotTracks(list(itrack, gtrack, atrack, grtrack,
dtrack), from = lim[1], to = lim[2])
#Change plot type to histogram
plotTracks(list(itrack, gtrack, atrack, grtrack,dtrack),
from = lim[1], to = lim[2], type = "histogram")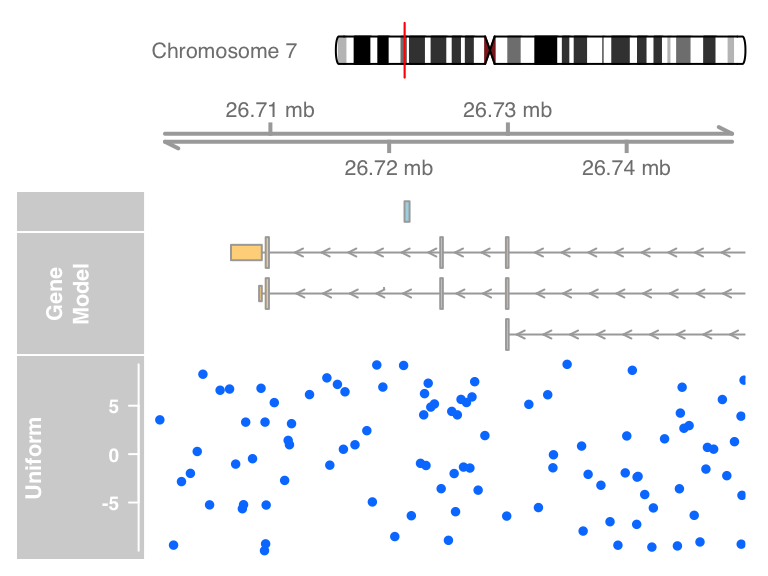
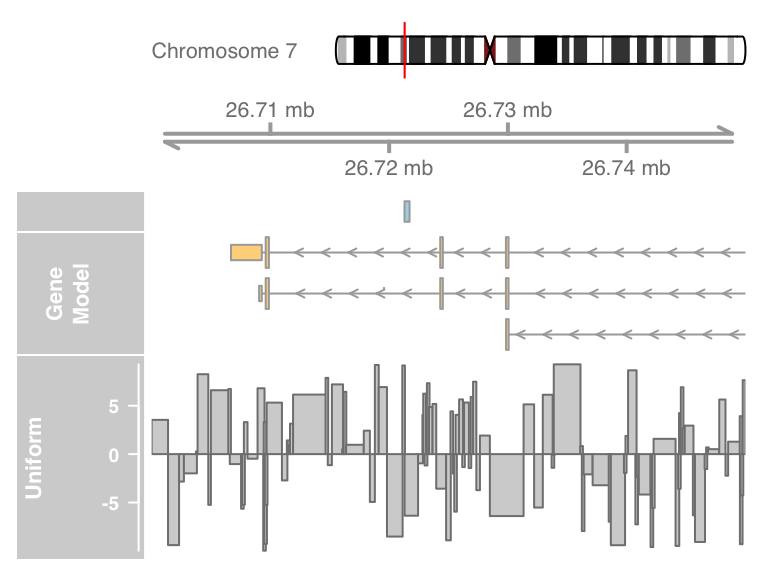
Such a visualization can be particularly helpful when displaying for instance the coverage of NGS reads along a chromosome, or to show the measurement values of mapped probes from a micro array experiment.Plotting parameters
setting parameters
#Annotation of transcript
#Change panel and title background color
grtrack <- GeneRegionTrack(geneModels, genome = gen,
chromosome = chr, name = "Gene Model",
transcriptAnnotation = "symbol",
background.panel = "#FFFEDB",
background.title = "darkblue")
plotTracks(list(itrack, gtrack, atrack, grtrack))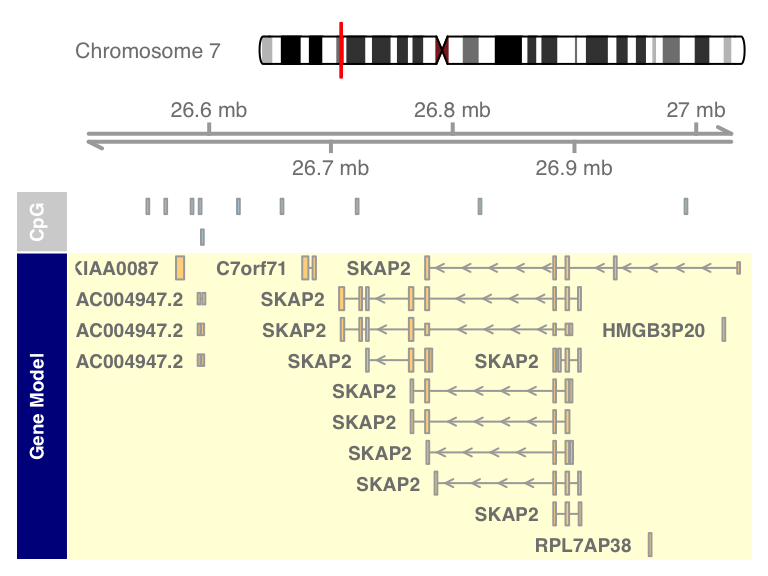
Plotting direction
By default all tracks will be plotted in a 5’ -> 3’ direction. It sometimes can be useful to actually show the data relative to the opposite strand.
plotTracks(list(itrack, gtrack, atrack, grtrack),
reverseStrand = TRUE)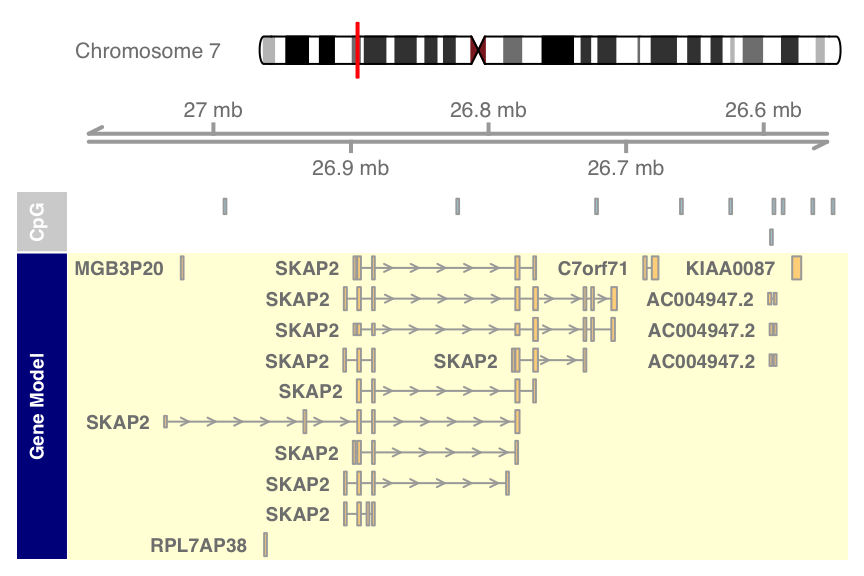
As you can see, the fact that the data has been plotted on the reverse strand is also reflected in the GenomeAxis track.Track classes
Several parameters can be used to change the appearance of the different tracks.
Display parameters for GenomeAxisTrack objects
Set the position of labels to below, show IDs, change color
axisTrack <- GenomeAxisTrack(range = IRanges(start = c(2000000,4000000),
end = c(3000000, 7000000),
names = rep("N-stretch", 2))
)
plotTracks(axisTrack, from = 1000000, to = 9000000,
labelPos = "below",showId=TRUE, col="red")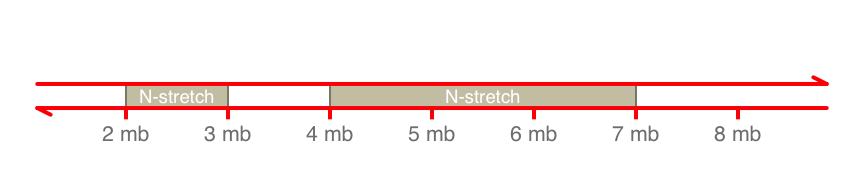
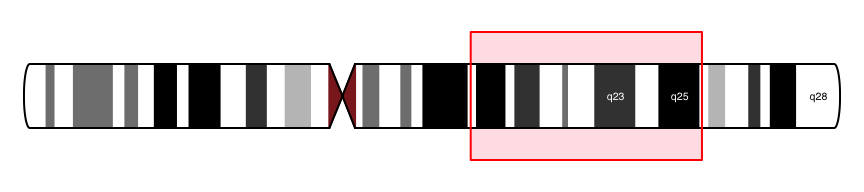
IdeogramTrack
#Ideogram
ideoTrack <- IdeogramTrack(genome = "hg19", chromosome = "chrX")
plotTracks(ideoTrack, from = 85000000, to = 129000000)
#Show chromosome band ID
plotTracks(ideoTrack, from = 85000000, to = 129000000,
showId = FALSE, showBandId = TRUE, cex.bands = 0.4)
DataTrack
Essentially they constitute run-length encoded numeric vectors or matrices associated to a particular genomic coordinate range. There can be multiple samples in a single data set, and the plotting method provides tools to incorporate sample group information.
Thus the starting point for creating DataTrack objects will always be a set of ranges, either in the form of an IRanges or GRanges object, or individually as start and end coordinates or widths. The second ingredient is a numeric vector of the same length as the number of ranges, or a numeric matrix with the same number of columns.
We will load our sample data from an GRanges object that comes as part of the Gviz package.
#Load data
data(twoGroups)
head(twoGroups)## GRanges with 6 ranges and 6 metadata columns:
## seqnames ranges strand | control control.1 control.2 treated
## |
## [1] chrX [ 1, 30] * | -8.96125989500433 -7.65790161676705 9.87956526689231 -5.84375557024032
## [2] chrX [ 42, 71] * | -4.2114706709981 4.6882571419701 -1.0533055011183 1.03083667811006
## [3] chrX [ 84, 113] * | 2.28711236733943 8.01326935179532 -7.1219984581694 -4.46718293242157
## [4] chrX [125, 154] * | 9.20983788557351 -6.23242623638362 8.59682233538479 -6.32041404955089
## [5] chrX [167, 196] * | 0.406841854564846 -7.05442394595593 -0.551973707042634 9.36362744309008
## [6] chrX [209, 238] * | 5.90989288408309 -5.10347711388022 1.56467542983592 -0.488725560717285
## treated.1 treated.2
##
## [1] 9.71352839842439 9.99328563921154
## [2] -6.77430204115808 0.593712376430631
## [3] -4.05887754634023 8.05319488979876
## [4] -1.56806231010705 3.5114610241726
## [5] -4.88056596834213 1.55288028530777
## [6] 6.99816173873842 -2.03484911937267
## ---
## seqlengths:
## chrX
## NA #Plot data track
dTrack <- DataTrack(twoGroups, name = "uniform")
plotTracks(dTrack)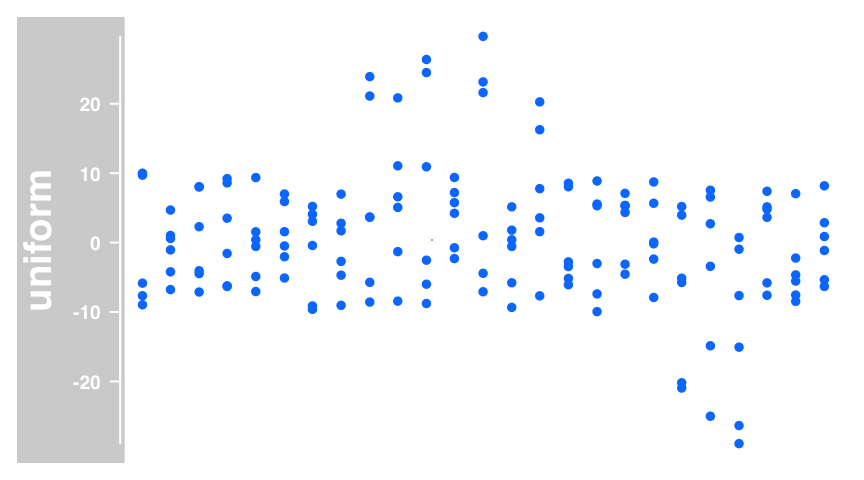
The default visualization for DataTrack is a dot plot.
The different plot types
The possible plot types are :
#dotplot
plotTracks(DataTrack(twoGroups, name = "p"), type="p")
#lines plot
plotTracks(DataTrack(twoGroups, name = "l"), type="l")
#line and dot plot
plotTracks(DataTrack(twoGroups, name = "b"), type="b")
#lines plot of average
plotTracks(DataTrack(twoGroups, name = "a"), type="a")
#histogram lines
plotTracks(DataTrack(twoGroups, name = "h"), type="h")
#histogram histogram (bar width equal to range with)
plotTracks(DataTrack(twoGroups, name = "histogram"), type="histogram")
#'polygon-type' plot relative to a baseline
plotTracks(DataTrack(twoGroups, name = "polygon"), type="polygon")
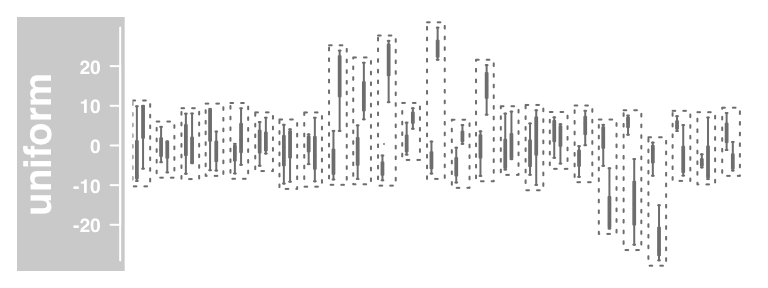
#box and whisker plot
plotTracks(DataTrack(twoGroups, name = "boxplot"), type="boxplot")
#false color image of the individual values
plotTracks(DataTrack(twoGroups, name = "heatmap"), type="heatmap")
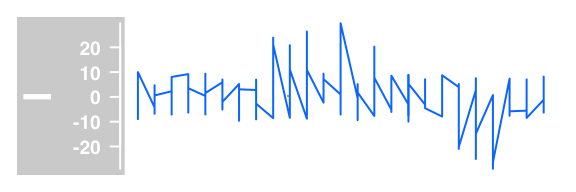
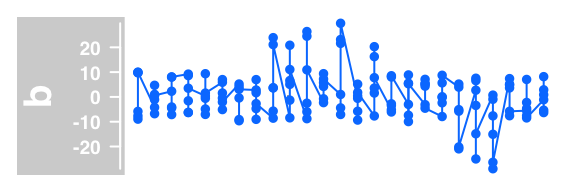
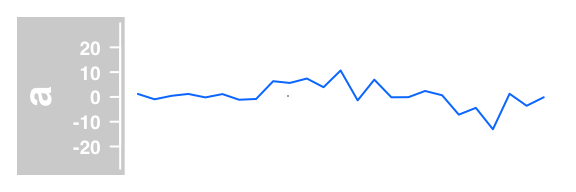
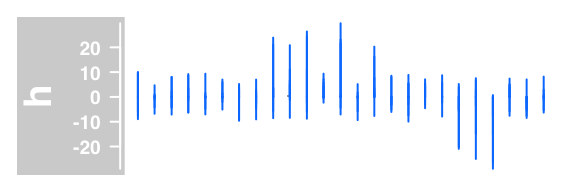
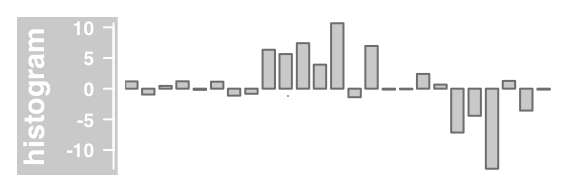
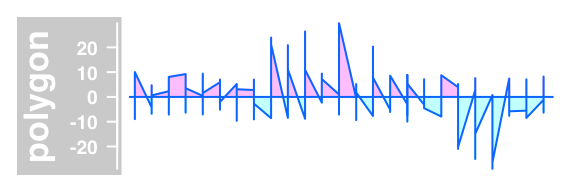
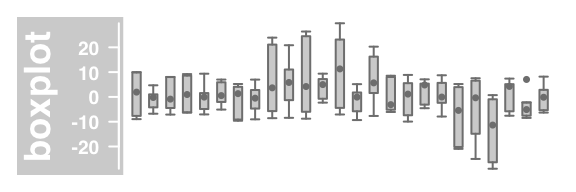
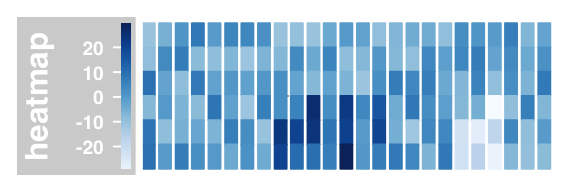
Example of DataTrack plots :
#Combine a boxplot with an average line and a data grid (g):
plotTracks(dTrack, type = c("boxplot", "a", "g"))
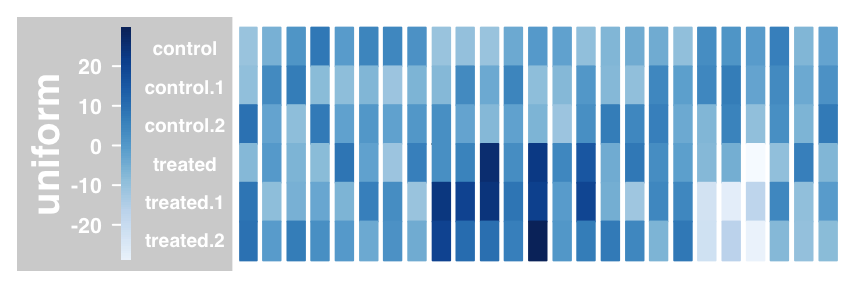
#Heatmap and show sample names
plotTracks(dTrack, type = c("heatmap"), showSampleNames = TRUE,
cex.sampleNames = 0.6)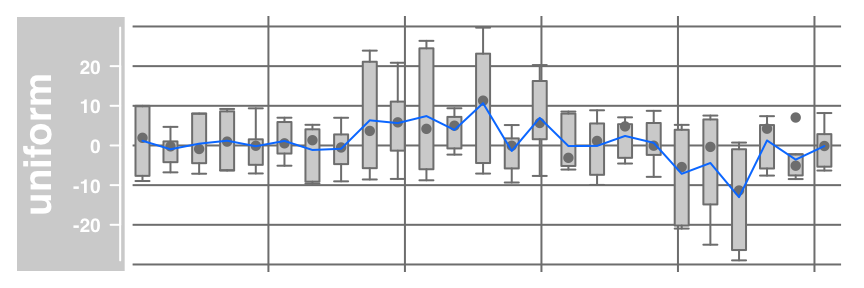
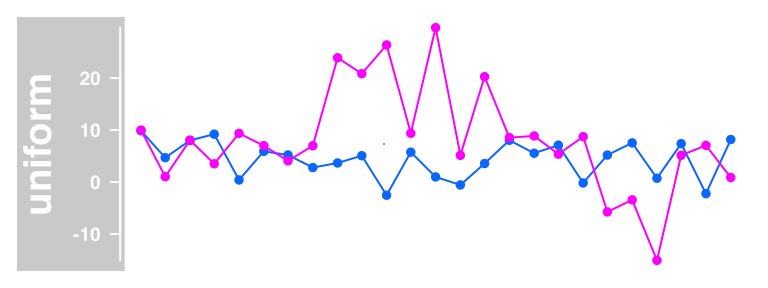
Data grouping
The individual samples can be grouped together using a factor variable.
plotTracks(dTrack, groups = rep(c("control", "treated"), each = 3),
type = c("a", "p"), legend=TRUE)
#Boxplot
plotTracks(dTrack, groups = rep(c("control", "treated"), each = 3),
type = "boxplot")
#Aggregate group. aggregation can be mean, median, extreme,
#sum, min and max
plotTracks(dTrack, groups = rep(c("control", "treated"),each = 3),
type = c("b"), aggregateGroups = TRUE,
aggregation = "max")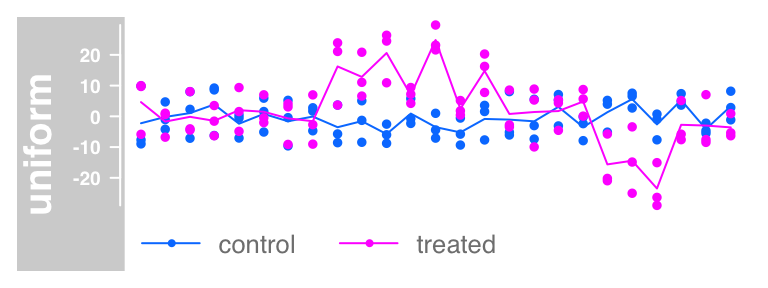
Building DataTrack objects from files
The DataTrack class supports the most common file types like wig, bigWig, bedGraph and bam files.
bgFile <- system.file("extdata/test.bedGraph", package = "Gviz")
dTrack2 <- DataTrack(range = bgFile, genome = "hg19",
type = "l", chromosome = "chr19", name = "bedGraph")
plotTracks(dTrack2)Note that the
Gviz package is using functionality from the rtracklayer package for most of the file import operationsThe real power of the file support in the Gviz package comes with streaming from indexed files. Only the relevant part of the data has to be loaded during the plotting operation, so the underlying data files may be quite large without decreasing the performance or causing too big of a memory footprint.
We will exemplify this feature here using a small bam file that is provided with the package. bam files contain alignments of sequences (typically from a next generation sequencing experiment) to a common reference. The most natural representation of such data in a DataTrack is to look at the alignment coverage at a given position only and to encode this in a single elementMetadata column.
bamFile <- system.file("extdata/test.bam", package = "Gviz")
dTrack4 <- DataTrack(range = bamFile, genome = "hg19",
type = "l", name = "Coverage", window = -1, chromosome = "chr1")
plotTracks(dTrack4, from = 189990000, to = 190000000)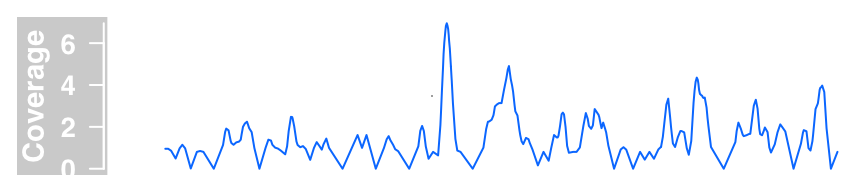
AnnotationTrack
Essentially they consist of one or several genomic ranges that can be grouped into composite annotation elements if needed. The necessary building blocks are the range coordinates, a chromosome and a genome identifier. Information can be passed to the function, either in the form of separate function arguments, as IRanges, GRanges or data.frame objects.
aTrack <- AnnotationTrack(start = c(10, 40, 120),
width = 15, chromosome = "chrX", strand = c("+","*", "-"),
id = c("Huey", "Dewey", "Louie"),
genome = "hg19", name = "foo")
plotTracks(aTrack)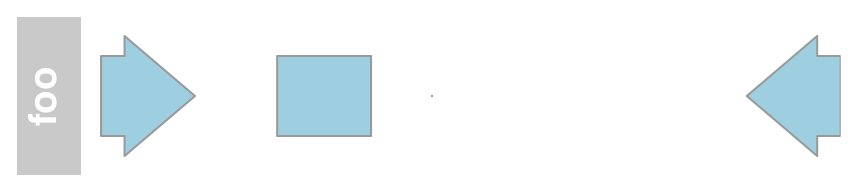
Building AnnotationTrack objects from files
The default import function reads the coordinates of all the sequence alignments from the bam file.
#Annotation track
aTrack2 <- AnnotationTrack(range = bamFile, genome = "hg19",
name = "Reads", chromosome = "chr1")
plotTracks(aTrack2, from = 189995000, to = 190000000)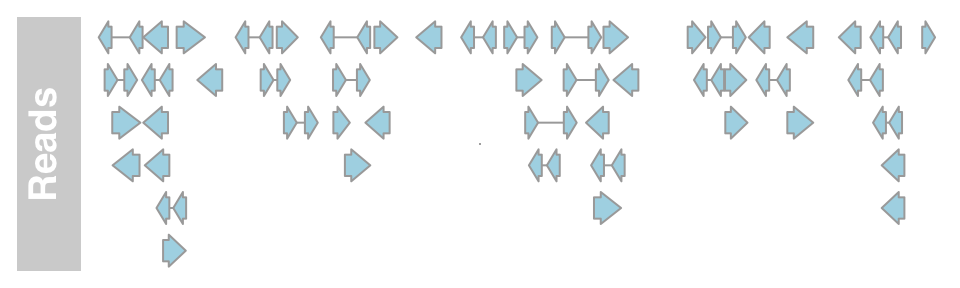
We can now plot both the DataTrack representation as well as the AnnotationTrack representation of the bam file together to prove that the underlying data are indeed identical.
plotTracks(list(dTrack4, aTrack2), from = 189990000,
to = 190000000)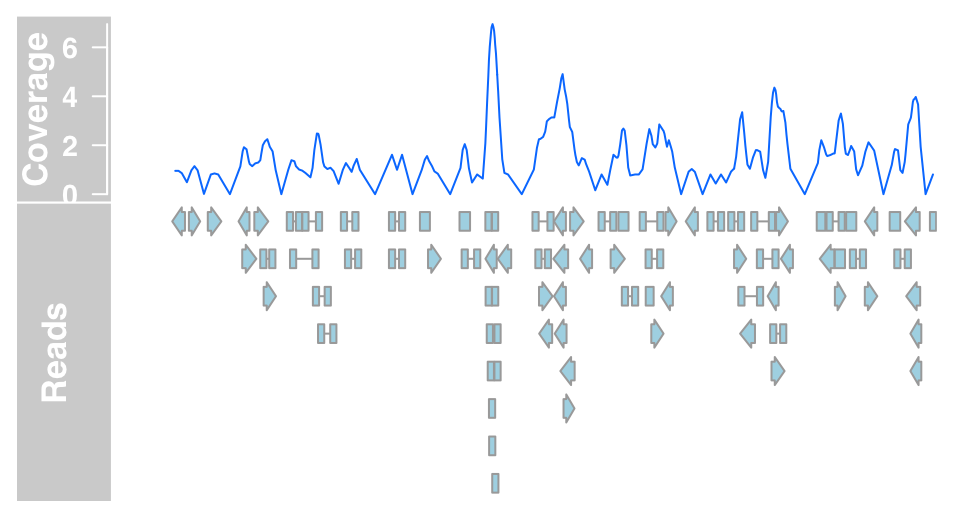
GeneRegionTrack
GeneRegionTrack objects are in principle very similar to AnnotationTrack objects. The only difference is that they are a little more gene/transcript centric. We need to pass start and end positions (or the width) of each annotation feature in the track and also supply the exon, transcript and gene identifiers for each item which will be used to create the transcript groupings.
A somewhat special case is to build a GeneRegionTrack object directly from one of the popular TranscriptDb objects, an option that is treated in more detail below.data(geneModels)
grtrack <- GeneRegionTrack(geneModels, genome = gen,
chromosome = chr, name = "foo",
transcriptAnnotation = "symbol")Building GeneRegionTrack objects from TranscriptDbs
The GenomicFeatures packages provides an elegant framework to download gene model information from online sources and to store it locally in a SQLite data base.
A nice bonus when building GeneRegionTracks from TranscriptDb objects is that we get additional information about coding and non-coding regions of the transcripts, i.e., coordinates of the 5’ and 3’ UTRs and of the CDS regions.
library(GenomicFeatures)
samplefile <- system.file("extdata", "UCSC_knownGene_sample.sqlite",
package = "GenomicFeatures")
txdb <- loadDb(samplefile)
txTr <- GeneRegionTrack(txdb, chromosome = "chr6", start = 300000, end = 350000)
#feature(txTr)
plotTracks(txTr)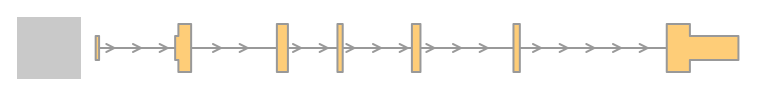
BiomartGeneRegionTrack
BiomartGeneRegionTrack class, provides a direct interface to the ENSEMBL Biomart service. We just enter a genome, chromosome and a start and end position on this chromosome, and the constructor function BiomartGeneRegionTrack will automatically contact ENSEMBL, fetch the necessary information and build the gene model on the fly.
Please note that you will need an internet connection for this to work, and that contacting Biomart can take a significant amount of time depending on usage and network trafic. biomTrack <- BiomartGeneRegionTrack(genome = "hg19",
chromosome = chr, start = 20000000, end = 21000000,
name = "ENSEMBL")
plotTracks(biomTrack, col.line = NULL, col = NULL)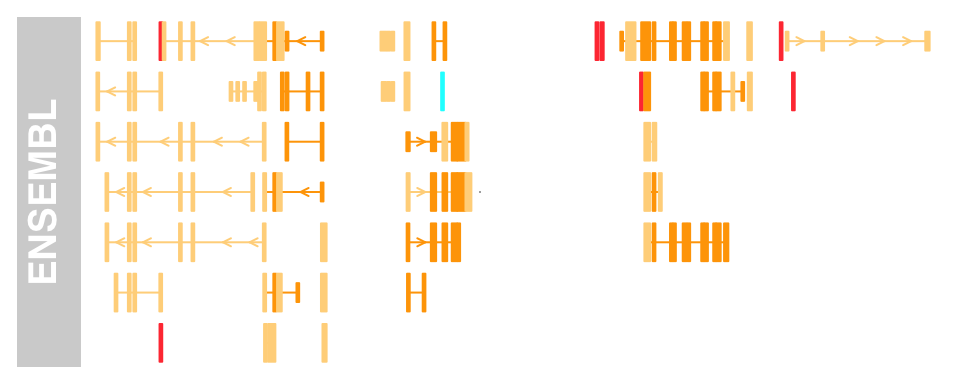
Sequence Track
library(BSgenome.Hsapiens.UCSC.hg19)
sTrack <- SequenceTrack(Hsapiens)
#sequence track : add 5'->3'
plotTracks(sTrack, chromosome = 1, from = 20000,to = 20050,
add53=TRUE)
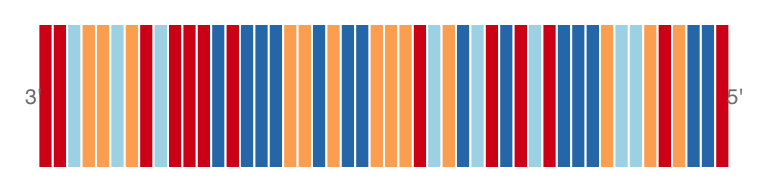
#The complement
plotTracks(sTrack, chromosome = 1, from = 20000,to = 20050,
add53=TRUE, complement = TRUE)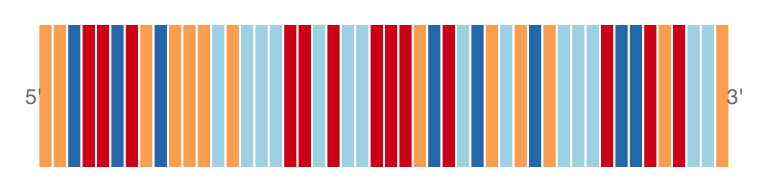
AlignmentsTrack
Plots of aligned sequences, typically from next generation sequencing experiments can be quite helpful, for instance when visually inspecting the validity of a called SNP. Those alignments are usually stored in BAM files.
RNAseq experiment
For this demonstration let’s use a small BAM file for which paired NGS reads have been mapped to an extract of the human hg19 genome. The data originate from an RNASeq experiment, and the alignments have been performed using the STAR aligner allowing for gaps. We also download some gene annotation data for that region from Biomart.
afrom=2960000
ato=3160000
#bam file
alTrack <- AlignmentsTrack(system.file(package = "Gviz", "extdata", "gapped.bam"), isPaired = TRUE)
bmt <- BiomartGeneRegionTrack(genome = "hg19", chromosome = "chr12",
start = afrom, end = ato, filter = list(with_ox_refseq_mrna = TRUE),
stacking = "dense")
plotTracks(c(bmt, alTrack), from = afrom, to = ato, chromosome = "chr12")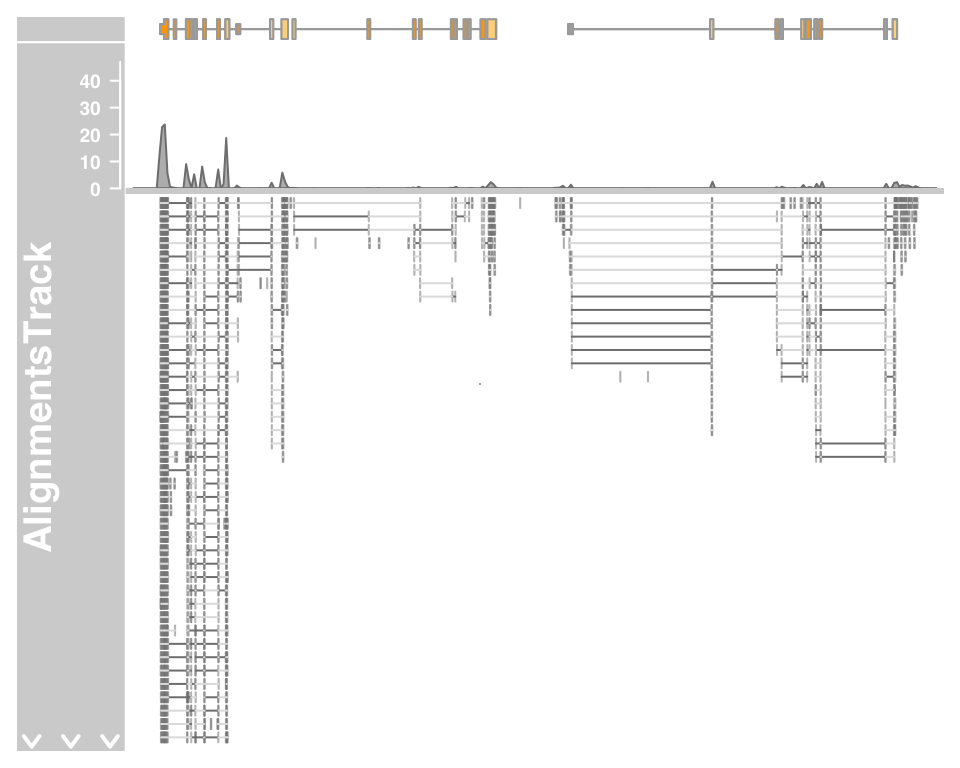
Now this already shows us the general layout of the track: on top we have a panel with the read coverage information in the form of a histogram, and below that a pile-up view of the individual reads. There is
only a certain amount of vertical space available for the plotting, and not the whole depth of the pile-up
can be displayed here. This fact is indicated by the white downward-pointing arrows in the title panel. We
could address this issue by playing around with the max.height, min.height or stackHeight display parameters which all control the height or the vertical spacing of the stacked reads. Or we could reduce the size of the coverage section by setting the coverageHeight or the minCoverageHeight parameters.plotTracks(c(bmt, alTrack), from = afrom, to = ato,
chromosome = "chr12", min.height = 0, coverageHeight = 0.08,
minCoverageHeight = 0)From that far out the pile-ups are not particularly useful, and we can turn those off by setting the type display parameter accordingly.
plotTracks(c(alTrack, bmt), from = afrom, to = ato, chromosome = "chr12", type = "coverage")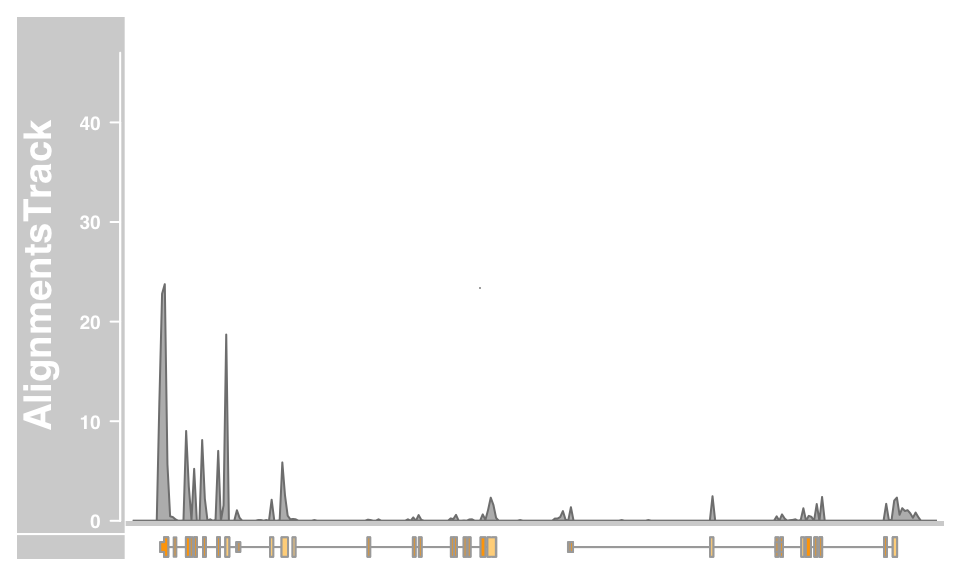
Let’s zoom in a bit further to check out the details of the pile-ups section:
plotTracks(c(bmt, alTrack), from = afrom + 12700,
to = afrom + 15200, chromosome = "chr12")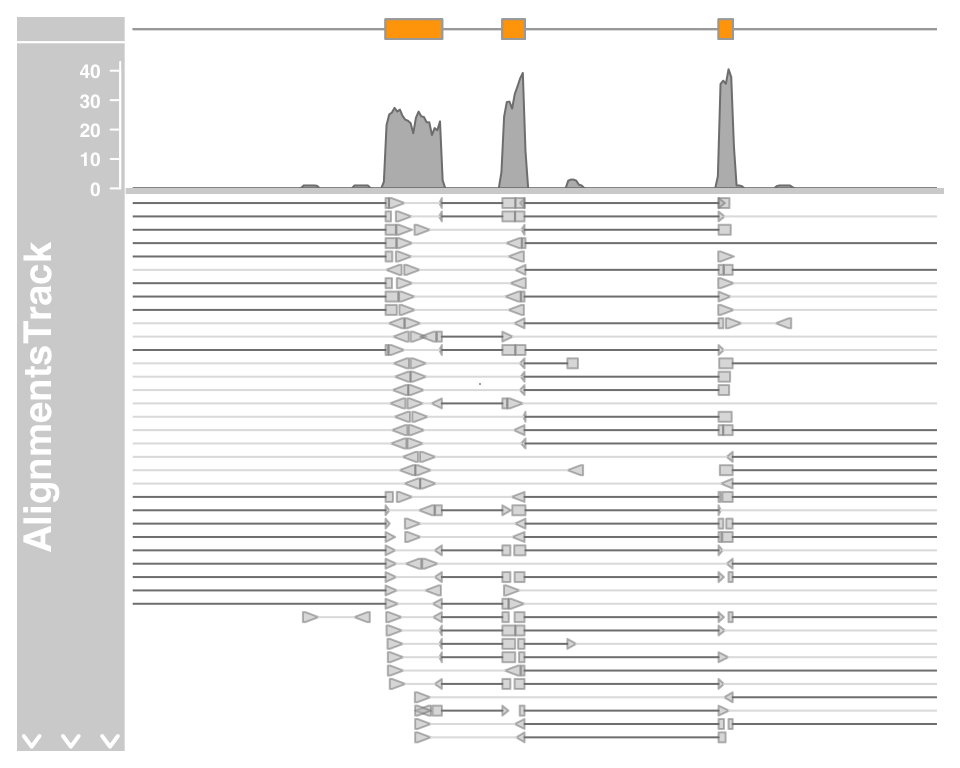
The direction of the individual reads is indicated by the arrow head, and read pairs are connect by a bright gray line. Gaps in the alignments are show by the connecting dark gray lines. On devices that support transparancy we can also see that some of the read pairs are actually overlapping. As mentioned before we can control whether the data should be treated as paired end or single end data by setting the isPaired argument in the constructor. Here is how we could take a look at the data in the same file, but in single end mode.
alTrack <- AlignmentsTrack(system.file(package = "Gviz",
"extdata", "gapped.bam"), isPaired = FALSE)
plotTracks(c(bmt, alTrack), from = afrom + 12700,
to = afrom + 15200, chromosome = "chr12")DNAseq experiment
To better show the features of the AlignmentsTrack for sequence variants we will load a different data set, this time from a whole genome DNASeq SNP calling experiment. Again the reference genome is hg19 and the alignments have been performed using Bowtie2.
We need to tell the AlignmentsTrack about the reference genome (sequenceTrack).
afrom <- 44945200
ato <- 44947200
alTrack <- AlignmentsTrack(system.file(package = "Gviz","extdata", "snps.bam"), isPaired = TRUE)
plotTracks(c(alTrack, sTrack), chromosome = "chr21", from = afrom,to = ato)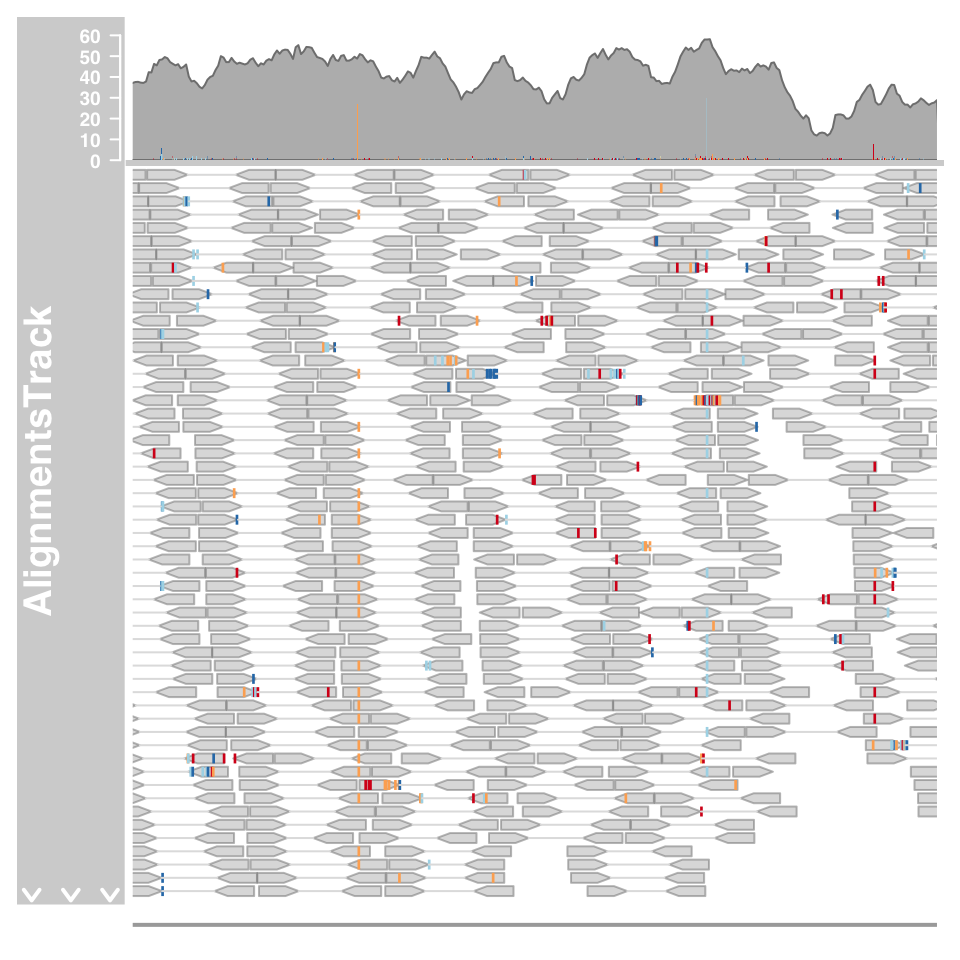
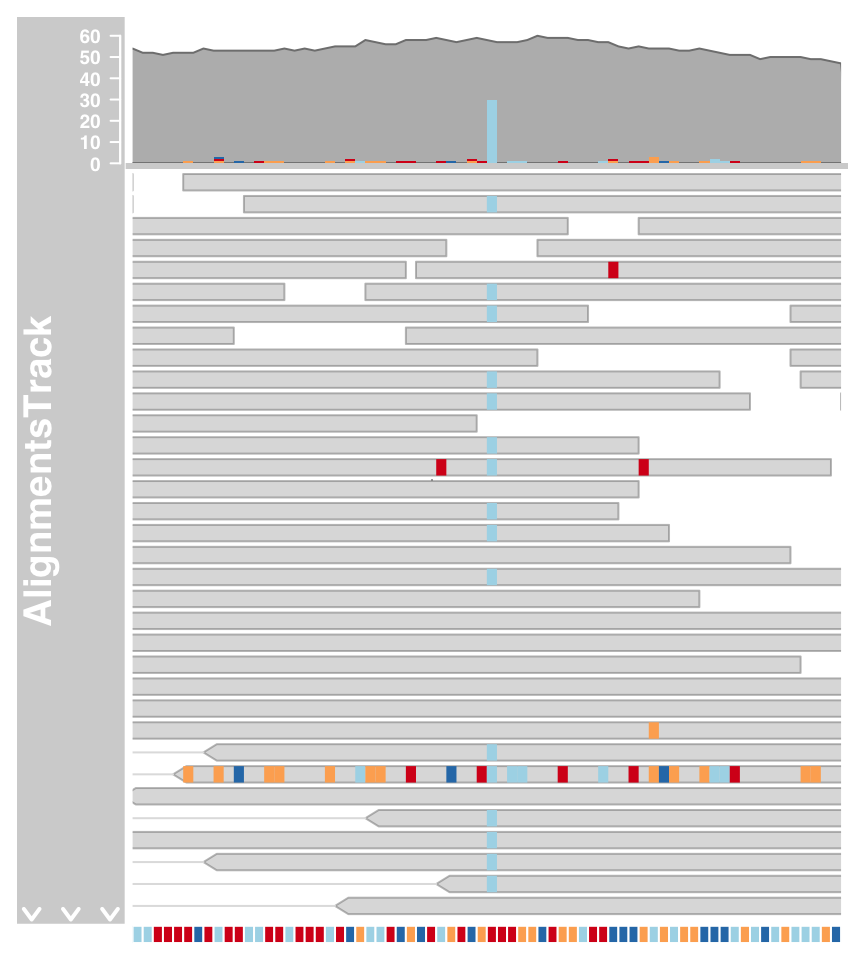
The mismatched bases are indicated on both the individual reads in the pileup section and also in the coverage plot in the form of a stacked histogram.When zooming in to one of the obvious heterozygous SNP positions we can reveal even more details.
#Zoom
plotTracks(c(alTrack, sTrack), chromosome = "chr21", from = 44946590, to = 44946660)
#show individual letters
plotTracks(c(alTrack, sTrack), chromosome = "chr21",
from = 44946590, to = 44946660, cex = 0.5, min.height = 8)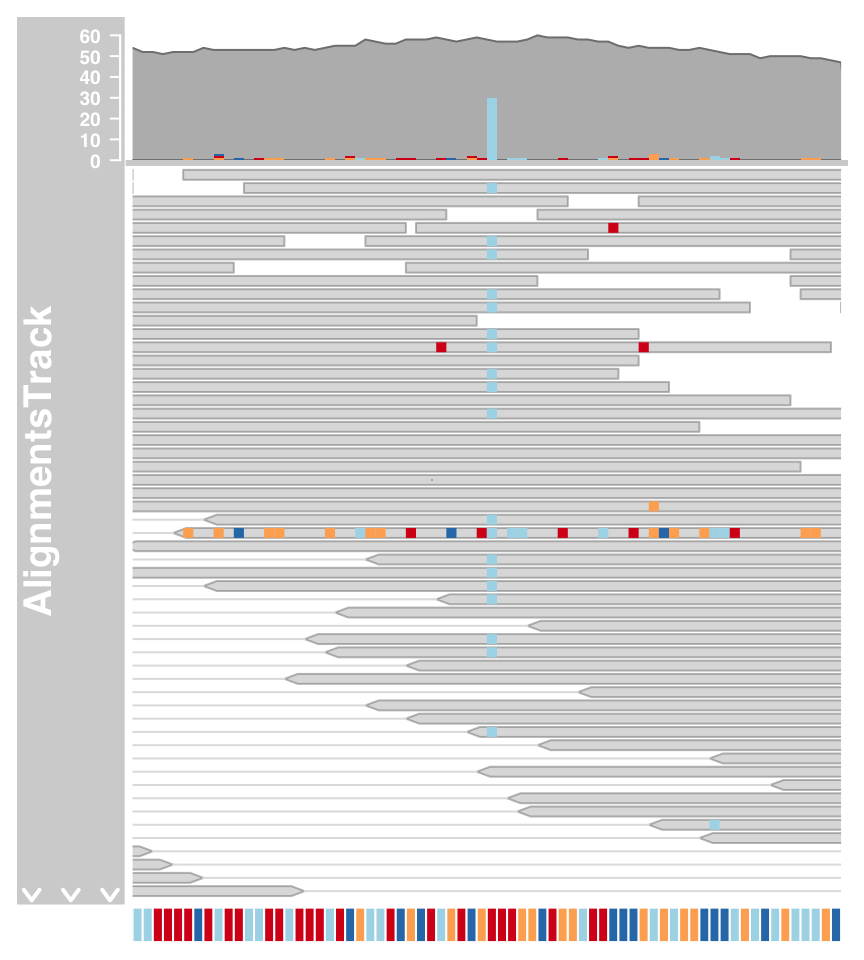
Track highlighting and overlays
Highlighting
#highlight
ht <- HighlightTrack(trackList = list(atrack, grtrack,dtrack),
start = c(26705000, 26720000), width = 7000, chromosome = 7)
plotTracks(list(itrack, gtrack, ht), from = lim[1], to = lim[2])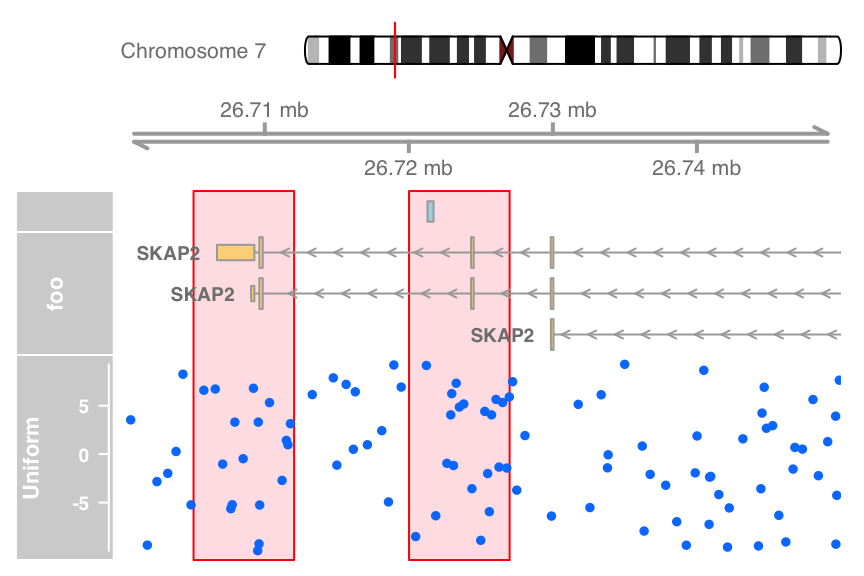
Overlays
For certain applications it can make sense to overlay multiple tracks on the same area of the plot. For the purpose of an instructive example we will generate a second DataTrack object and combine it with the existing one from the second chapter.
#create data
dat <- runif(100, min = -2, max = 22)
dtrack2 <- DataTrack(data = dat, start = coords[-length(coords)],
end = coords[-1], chromosome = chr, genome = gen,
name = "Uniform2", groups = factor("sample 2",levels = c("sample 1", "sample 2")),
legend = TRUE)
displayPars(dtrack) <- list(groups = factor("sample 1",levels = c("sample 1", "sample 2")), legend = TRUE)
ot <- OverlayTrack(trackList = list(dtrack2, dtrack))
ylims <- extendrange(range(c(values(dtrack), values(dtrack2))))
plotTracks(list(itrack, gtrack, ot), from = lim[1], to = lim[2], ylim = ylims, type = c("smooth", "p"))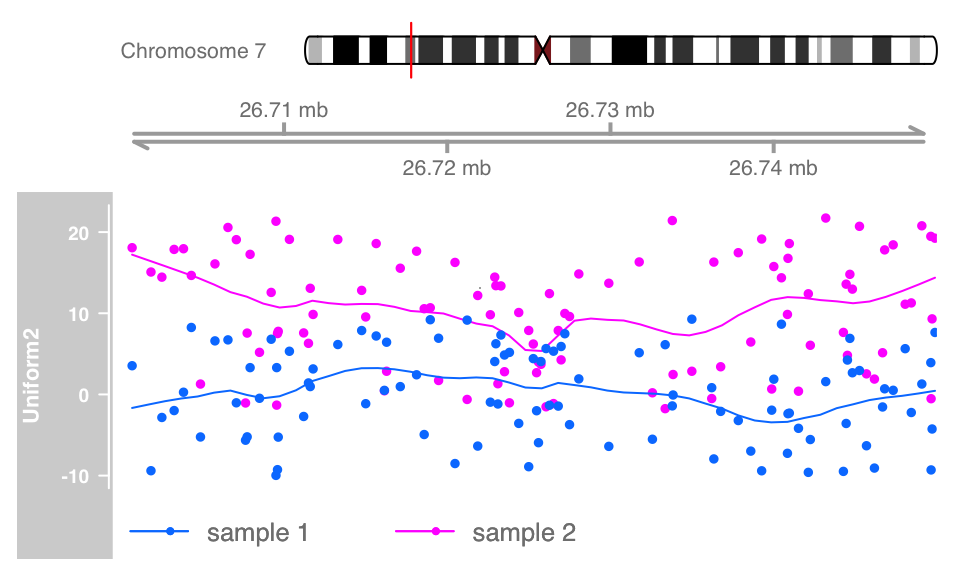
On devices that support it, alpha blending can be a useful tool to tease out even more information out of track overlays, at least when comparing just a small number of samples. The resulting transparency effectively eliminates the problem of overplotting. The following example will only work if this vignette has been built on a system with alpha blending support.
displayPars(dtrack) <- list(alpha.title = 1, alpha = 0.5)
displayPars(dtrack2) <- list(alpha.title = 1, alpha = 0.5)
ot <- OverlayTrack(trackList = list(dtrack, dtrack2))
plotTracks(list(itrack, gtrack, ot), from = lim[1],
to = lim[2], ylim = ylims, type = c("hist"), window = 30)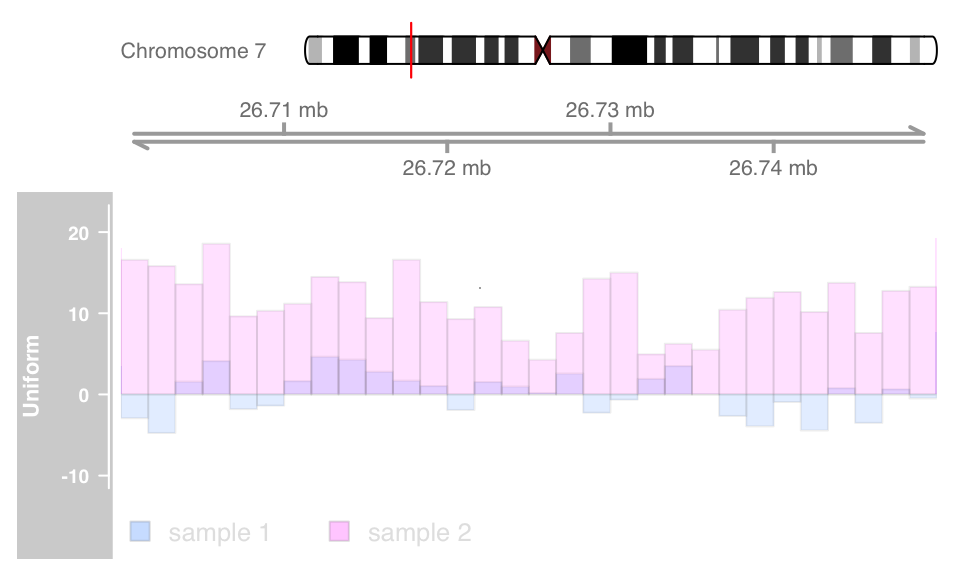
Licence
Show me some love with the like buttons below... Thank you and please don't forget to share and comment below!!
Montrez-moi un peu d'amour avec les like ci-dessous ... Merci et n'oubliez pas, s'il vous plaît, de partager et de commenter ci-dessous!
Recommended for You!
Recommended for you
This section contains the best data science and self-development resources to help you on your path.
Books - Data Science
Our Books
- Practical Guide to Cluster Analysis in R by A. Kassambara (Datanovia)
- Practical Guide To Principal Component Methods in R by A. Kassambara (Datanovia)
- Machine Learning Essentials: Practical Guide in R by A. Kassambara (Datanovia)
- R Graphics Essentials for Great Data Visualization by A. Kassambara (Datanovia)
- GGPlot2 Essentials for Great Data Visualization in R by A. Kassambara (Datanovia)
- Network Analysis and Visualization in R by A. Kassambara (Datanovia)
- Practical Statistics in R for Comparing Groups: Numerical Variables by A. Kassambara (Datanovia)
- Inter-Rater Reliability Essentials: Practical Guide in R by A. Kassambara (Datanovia)
Others
- R for Data Science: Import, Tidy, Transform, Visualize, and Model Data by Hadley Wickham & Garrett Grolemund
- Hands-On Machine Learning with Scikit-Learn, Keras, and TensorFlow: Concepts, Tools, and Techniques to Build Intelligent Systems by Aurelien Géron
- Practical Statistics for Data Scientists: 50 Essential Concepts by Peter Bruce & Andrew Bruce
- Hands-On Programming with R: Write Your Own Functions And Simulations by Garrett Grolemund & Hadley Wickham
- An Introduction to Statistical Learning: with Applications in R by Gareth James et al.
- Deep Learning with R by François Chollet & J.J. Allaire
- Deep Learning with Python by François Chollet
Click to follow us on Facebook :
Comment this article by clicking on "Discussion" button (top-right position of this page)







