Principal Component Analysis in R: prcomp vs princomp
This R tutorial describes how to perform a Principal Component Analysis (PCA) using the built-in R functions prcomp() and princomp(). You will learn how to predict new individuals and variables coordinates using PCA. We’ll also provide the theory behind PCA results.
Learn more about the basics and the interpretation of principal component analysis in our previous article: PCA - Principal Component Analysis Essentials.

Contents:
Related Book:

Practical Guide to Principal Component Methods in R
General methods for principal component analysis
There are two general methods to perform PCA in R :
- Spectral decomposition which examines the covariances / correlations between variables
- Singular value decomposition which examines the covariances / correlations between individuals
The function princomp() uses the spectral decomposition approach. The functions prcomp() and PCA()[FactoMineR] use the singular value decomposition (SVD).
According to the R help, SVD has slightly better numerical accuracy. Therefore, the function prcomp() is preferred compared to princomp().
prcomp() and princomp() functions
The simplified format of these 2 functions are :
prcomp(x, scale = FALSE)
princomp(x, cor = FALSE, scores = TRUE)- Arguments for prcomp():
x: a numeric matrix or data framescale: a logical value indicating whether the variables should be scaled to have unit variance before the analysis takes place
- Arguments for princomp():
x: a numeric matrix or data framecor: a logical value. If TRUE, the data will be centered and scaled before the analysisscores: a logical value. If TRUE, the coordinates on each principal component are calculated
The elements of the outputs returned by the functions prcomp() and princomp() includes :
| prcomp() name | princomp() name | Description |
|---|---|---|
| sdev | sdev | the standard deviations of the principal components |
| rotation | loadings | the matrix of variable loadings (columns are eigenvectors) |
| center | center | the variable means (means that were substracted) |
| scale | scale | the variable standard deviations (the scaling applied to each variable ) |
| x | scores | The coordinates of the individuals (observations) on the principal components. |
In the following sections, we’ll focus only on the function prcomp()
Package for PCA visualization
We’ll use the factoextra R package to create a ggplot2-based elegant visualization.
- You can install it from CRAN:
install.packages("factoextra")- Or, install the latest developmental version from github:
if(!require(devtools)) install.packages("devtools")
devtools::install_github("kassambara/factoextra")- Load factoextra as follow:
library(factoextra)Demo data sets
We’ll use the data sets decathlon2 [in factoextra], which has been already described at: PCA - Data format.
Briefly, it contains:
- Active individuals (rows 1 to 23) and active variables (columns 1 to 10), which are used to perform the principal component analysis
- Supplementary individuals (rows 24 to 27) and supplementary variables (columns 11 to 13), which coordinates will be predicted using the PCA information and parameters obtained with active individuals/variables.
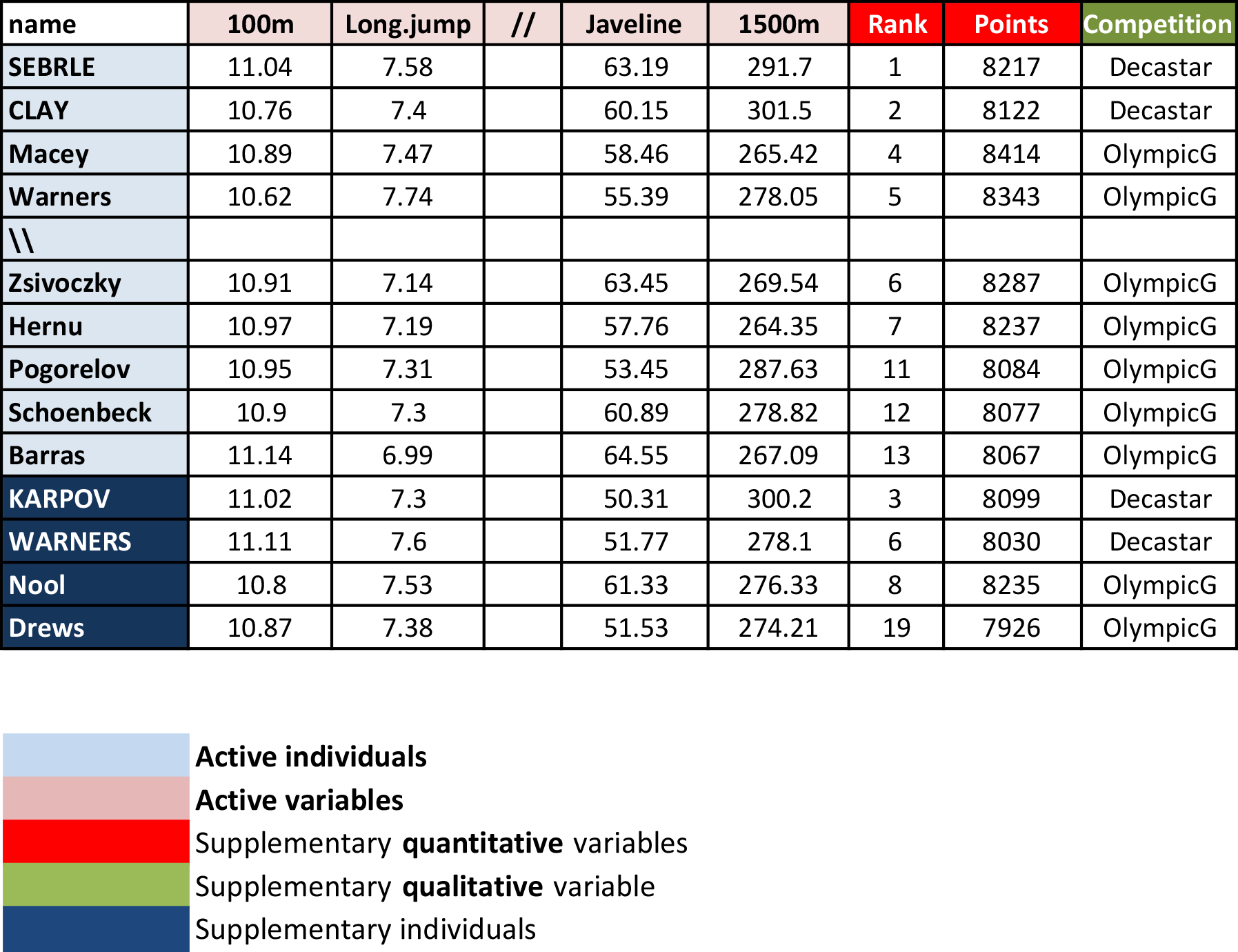
Load the data and extract only active individuals and variables:
library("factoextra")
data(decathlon2)
decathlon2.active <- decathlon2[1:23, 1:10]
head(decathlon2.active[, 1:6])## X100m Long.jump Shot.put High.jump X400m X110m.hurdle
## SEBRLE 11.0 7.58 14.8 2.07 49.8 14.7
## CLAY 10.8 7.40 14.3 1.86 49.4 14.1
## BERNARD 11.0 7.23 14.2 1.92 48.9 15.0
## YURKOV 11.3 7.09 15.2 2.10 50.4 15.3
## ZSIVOCZKY 11.1 7.30 13.5 2.01 48.6 14.2
## McMULLEN 10.8 7.31 13.8 2.13 49.9 14.4Compute PCA in R using prcomp()
In this section we’ll provide an easy-to-use R code to compute and visualize PCA in R using the prcomp() function and the factoextra package.
- Load factoextra for visualization
library(factoextra)- Compute PCA
res.pca <- prcomp(decathlon2.active, scale = TRUE)- Visualize eigenvalues (scree plot). Show the percentage of variances explained by each principal component.
fviz_eig(res.pca)
- Graph of individuals. Individuals with a similar profile are grouped together.
fviz_pca_ind(res.pca,
col.ind = "cos2", # Color by the quality of representation
gradient.cols = c("#00AFBB", "#E7B800", "#FC4E07"),
repel = TRUE # Avoid text overlapping
)
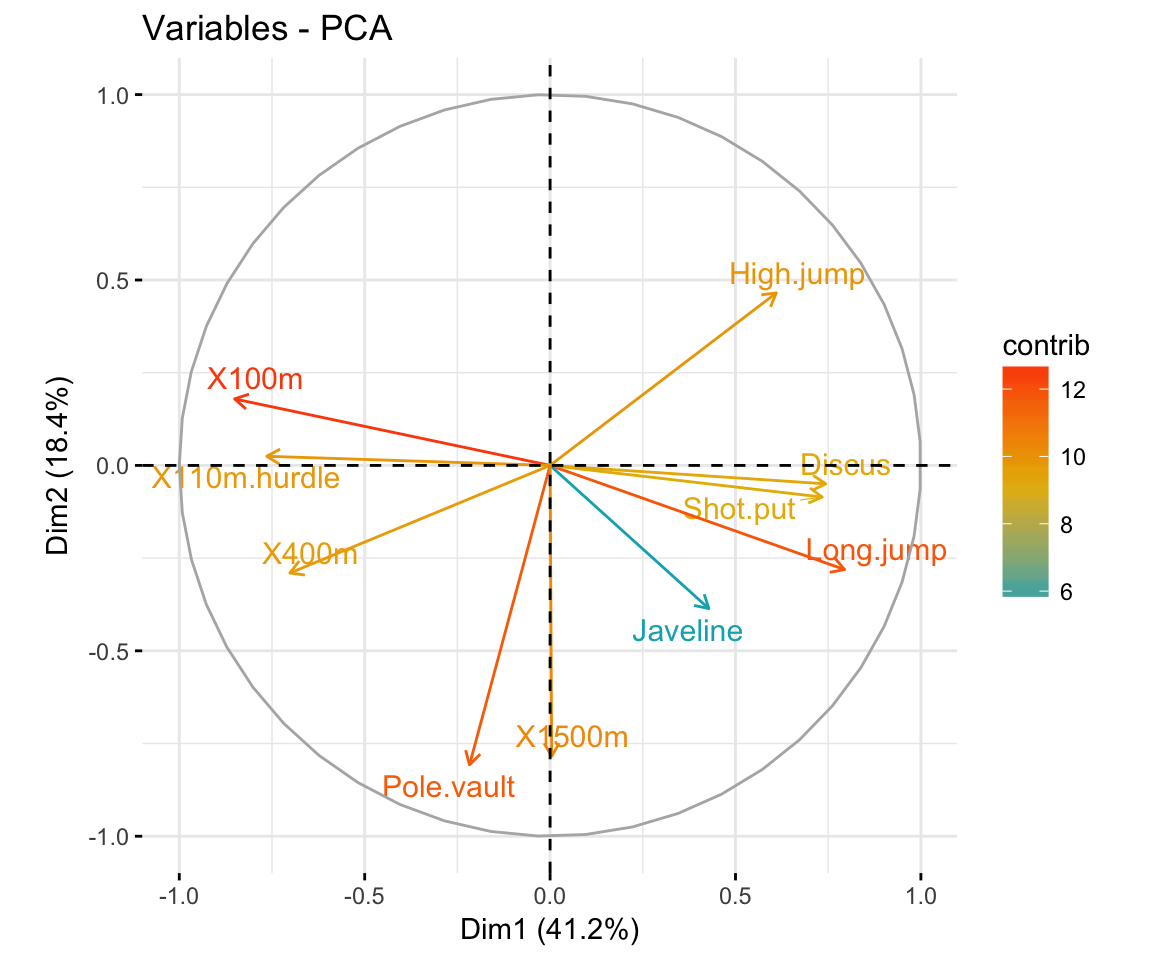
- Graph of variables. Positive correlated variables point to the same side of the plot. Negative correlated variables point to opposite sides of the graph.
fviz_pca_var(res.pca,
col.var = "contrib", # Color by contributions to the PC
gradient.cols = c("#00AFBB", "#E7B800", "#FC4E07"),
repel = TRUE # Avoid text overlapping
)
- Biplot of individuals and variables
fviz_pca_biplot(res.pca, repel = TRUE,
col.var = "#2E9FDF", # Variables color
col.ind = "#696969" # Individuals color
)
Access to the PCA results
library(factoextra)
# Eigenvalues
eig.val <- get_eigenvalue(res.pca)
eig.val
# Results for Variables
res.var <- get_pca_var(res.pca)
res.var$coord # Coordinates
res.var$contrib # Contributions to the PCs
res.var$cos2 # Quality of representation
# Results for individuals
res.ind <- get_pca_ind(res.pca)
res.ind$coord # Coordinates
res.ind$contrib # Contributions to the PCs
res.ind$cos2 # Quality of representation Predict using PCA
In this section, we’ll show how to predict the coordinates of supplementary individuals and variables using only the information provided by the previously performed PCA.
Supplementary individuals
- Data: rows 24 to 27 and columns 1 to to 10 [in decathlon2 data sets]. The new data must contain columns (variables) with the same names and in the same order as the active data used to compute PCA.
# Data for the supplementary individuals
ind.sup <- decathlon2[24:27, 1:10]
ind.sup[, 1:6]## X100m Long.jump Shot.put High.jump X400m X110m.hurdle
## KARPOV 11.0 7.30 14.8 2.04 48.4 14.1
## WARNERS 11.1 7.60 14.3 1.98 48.7 14.2
## Nool 10.8 7.53 14.3 1.88 48.8 14.8
## Drews 10.9 7.38 13.1 1.88 48.5 14.0- Predict the coordinates of new individuals data. Use the R base function predict():
ind.sup.coord <- predict(res.pca, newdata = ind.sup)
ind.sup.coord[, 1:4]## PC1 PC2 PC3 PC4
## KARPOV 0.777 -0.762 1.597 1.686
## WARNERS -0.378 0.119 1.701 -0.691
## Nool -0.547 -1.934 0.472 -2.228
## Drews -1.085 -0.017 2.982 -1.501- Graph of individuals including the supplementary individuals:
# Plot of active individuals
p <- fviz_pca_ind(res.pca, repel = TRUE)
# Add supplementary individuals
fviz_add(p, ind.sup.coord, color ="blue")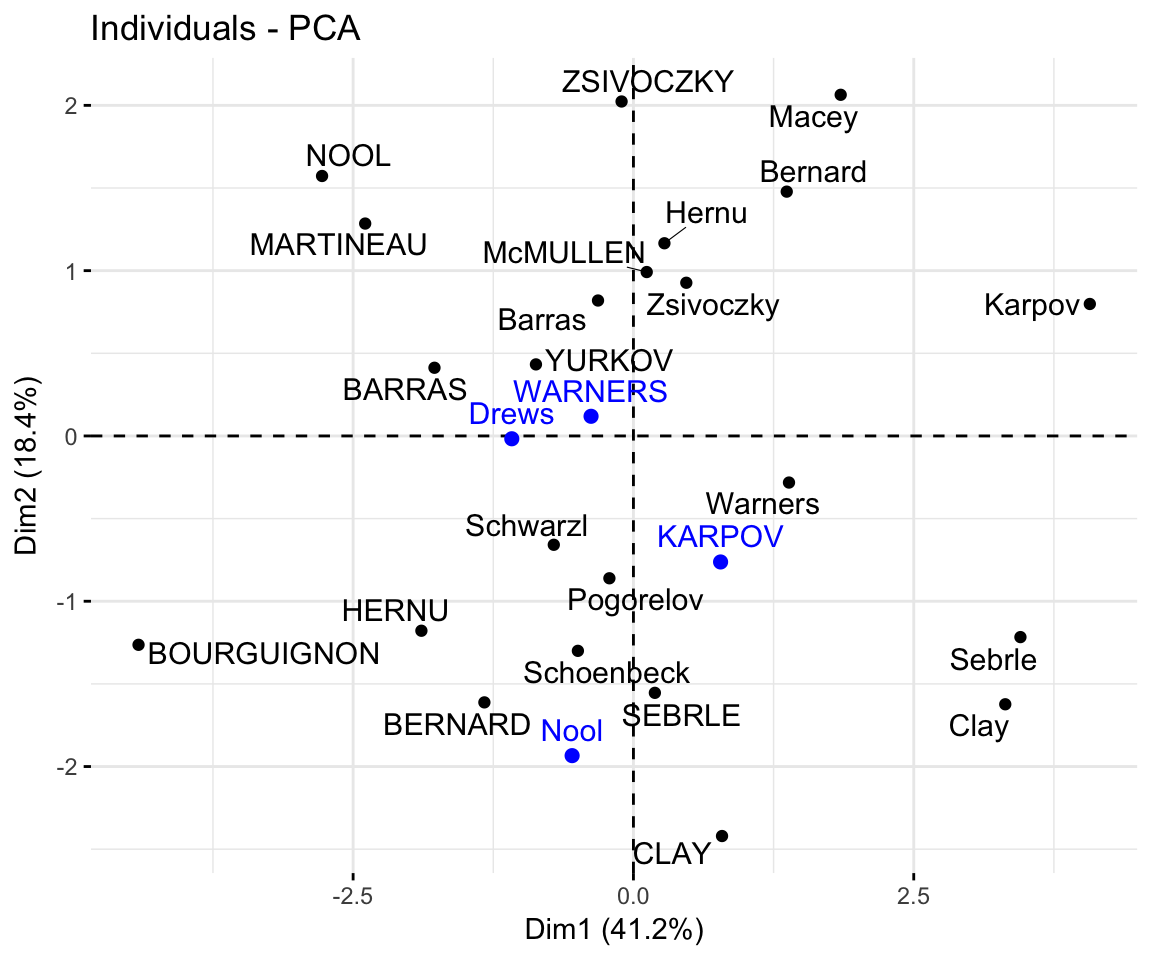
The predicted coordinates of individuals can be manually calculated as follow:
- Center and scale the new individuals data using the center and the scale of the PCA
- Calculate the predicted coordinates by multiplying the scaled values with the eigenvectors (loadings) of the principal components.
The R code below can be used :
# Centering and scaling the supplementary individuals
ind.scaled <- scale(ind.sup,
center = res.pca$center,
scale = res.pca$scale)
# Coordinates of the individividuals
coord_func <- function(ind, loadings){
r <- loadings*ind
apply(r, 2, sum)
}
pca.loadings <- res.pca$rotation
ind.sup.coord <- t(apply(ind.scaled, 1, coord_func, pca.loadings ))
ind.sup.coord[, 1:4]## PC1 PC2 PC3 PC4
## KARPOV 0.777 -0.762 1.597 1.686
## WARNERS -0.378 0.119 1.701 -0.691
## Nool -0.547 -1.934 0.472 -2.228
## Drews -1.085 -0.017 2.982 -1.501Supplementary variables
Qualitative / categorical variables
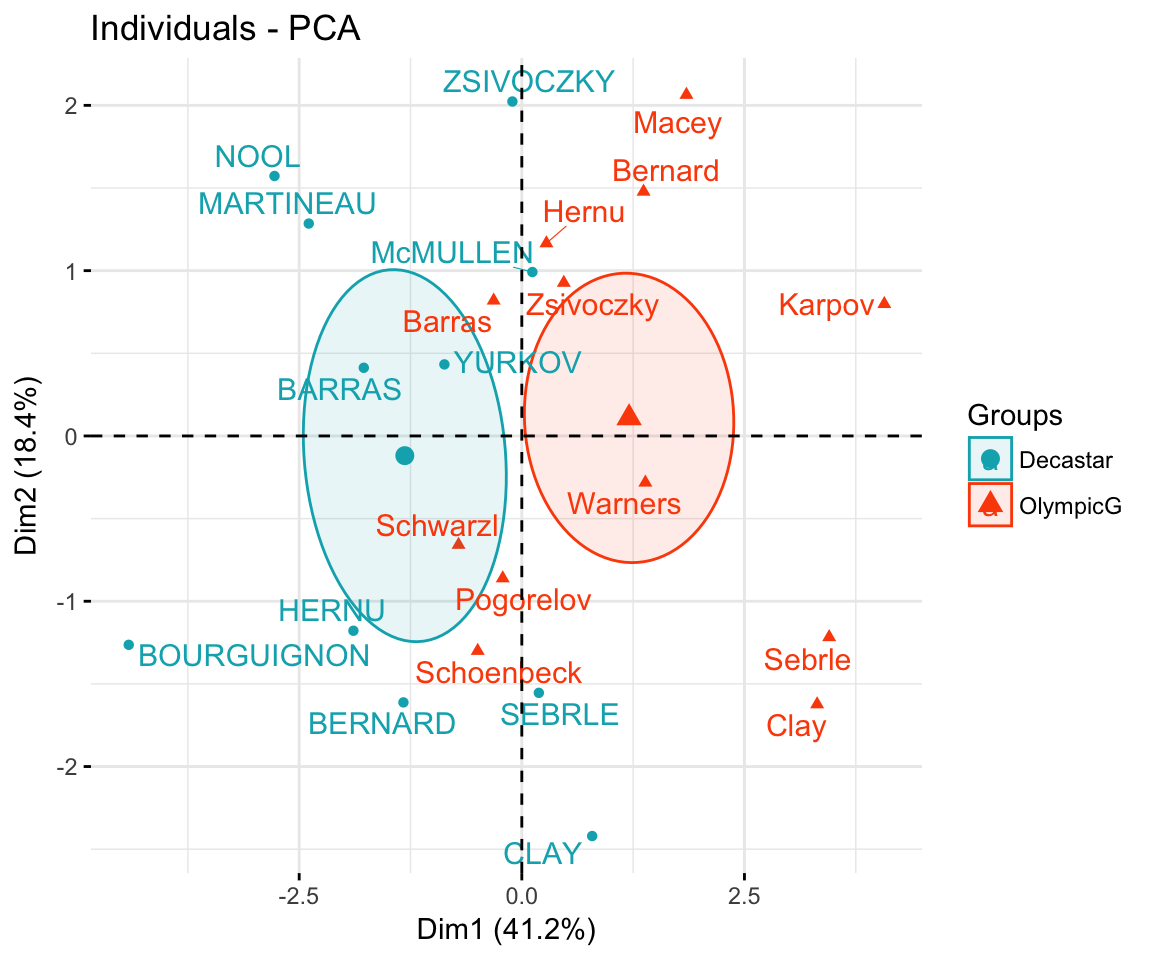
The data sets decathlon2 contain a supplementary qualitative variable at columns 13 corresponding to the type of competitions.
Qualitative / categorical variables can be used to color individuals by groups. The grouping variable should be of same length as the number of active individuals (here 23).
groups <- as.factor(decathlon2$Competition[1:23])
fviz_pca_ind(res.pca,
col.ind = groups, # color by groups
palette = c("#00AFBB", "#FC4E07"),
addEllipses = TRUE, # Concentration ellipses
ellipse.type = "confidence",
legend.title = "Groups",
repel = TRUE
)
Calculate the coordinates for the levels of grouping variables. The coordinates for a given group is calculated as the mean coordinates of the individuals in the group.
library(magrittr) # for pipe %>%
library(dplyr) # everything else
# 1. Individual coordinates
res.ind <- get_pca_ind(res.pca)
# 2. Coordinate of groups
coord.groups <- res.ind$coord %>%
as_data_frame() %>%
select(Dim.1, Dim.2) %>%
mutate(competition = groups) %>%
group_by(competition) %>%
summarise(
Dim.1 = mean(Dim.1),
Dim.2 = mean(Dim.2)
)
coord.groups## # A tibble: 2 x 3
## competition Dim.1 Dim.2
##
## 1 Decastar -1.31 -0.119
## 2 OlympicG 1.20 0.109 Quantitative variables
Data: columns 11:12. Should be of same length as the number of active individuals (here 23)
quanti.sup <- decathlon2[1:23, 11:12, drop = FALSE]
head(quanti.sup)## Rank Points
## SEBRLE 1 8217
## CLAY 2 8122
## BERNARD 4 8067
## YURKOV 5 8036
## ZSIVOCZKY 7 8004
## McMULLEN 8 7995The coordinates of a given quantitative variable are calculated as the correlation between the quantitative variables and the principal components.
# Predict coordinates and compute cos2
quanti.coord <- cor(quanti.sup, res.pca$x)
quanti.cos2 <- quanti.coord^2
# Graph of variables including supplementary variables
p <- fviz_pca_var(res.pca)
fviz_add(p, quanti.coord, color ="blue", geom="arrow")
Theory behind PCA results
PCA results for variables
Here we’ll show how to calculate the PCA results for variables: coordinates, cos2 and contributions:
var.coord= loadings * the component standard deviationsvar.cos2= var.coord^2var.contrib. The contribution of a variable to a given principal component is (in percentage) : (var.cos2 * 100) / (total cos2 of the component)
# Helper function
#::::::::::::::::::::::::::::::::::::::::
var_coord_func <- function(loadings, comp.sdev){
loadings*comp.sdev
}
# Compute Coordinates
#::::::::::::::::::::::::::::::::::::::::
loadings <- res.pca$rotation
sdev <- res.pca$sdev
var.coord <- t(apply(loadings, 1, var_coord_func, sdev))
head(var.coord[, 1:4])## PC1 PC2 PC3 PC4
## X100m -0.851 0.1794 -0.302 0.0336
## Long.jump 0.794 -0.2809 0.191 -0.1154
## Shot.put 0.734 -0.0854 -0.518 0.1285
## High.jump 0.610 0.4652 -0.330 0.1446
## X400m -0.702 -0.2902 -0.284 0.4308
## X110m.hurdle -0.764 0.0247 -0.449 -0.0169# Compute Cos2
#::::::::::::::::::::::::::::::::::::::::
var.cos2 <- var.coord^2
head(var.cos2[, 1:4])## PC1 PC2 PC3 PC4
## X100m 0.724 0.032184 0.0909 0.001127
## Long.jump 0.631 0.078881 0.0363 0.013315
## Shot.put 0.539 0.007294 0.2679 0.016504
## High.jump 0.372 0.216424 0.1090 0.020895
## X400m 0.492 0.084203 0.0804 0.185611
## X110m.hurdle 0.584 0.000612 0.2015 0.000285# Compute contributions
#::::::::::::::::::::::::::::::::::::::::
comp.cos2 <- apply(var.cos2, 2, sum)
contrib <- function(var.cos2, comp.cos2){var.cos2*100/comp.cos2}
var.contrib <- t(apply(var.cos2,1, contrib, comp.cos2))
head(var.contrib[, 1:4])## PC1 PC2 PC3 PC4
## X100m 17.54 1.7505 7.34 0.1376
## Long.jump 15.29 4.2904 2.93 1.6249
## Shot.put 13.06 0.3967 21.62 2.0141
## High.jump 9.02 11.7716 8.79 2.5499
## X400m 11.94 4.5799 6.49 22.6509
## X110m.hurdle 14.16 0.0333 16.26 0.0348PCA results for individuals
ind.coord= res.pca$x- Cos2 of individuals. Two steps:
- Calculate the square distance between each individual and the PCA center of gravity: d2 = [(var1_ind_i - mean_var1)/sd_var1]^2 + …+ [(var10_ind_i - mean_var10)/sd_var10]^2 + …+..
- Calculate the cos2 as ind.coord^2/d2
- Contributions of individuals to the principal components: 100 * (1 / number_of_individuals)*(ind.coord^2 / comp_sdev^2). Note that the sum of all the contributions per column is 100
# Coordinates of individuals
#::::::::::::::::::::::::::::::::::
ind.coord <- res.pca$x
head(ind.coord[, 1:4])## PC1 PC2 PC3 PC4
## SEBRLE 0.191 -1.554 -0.628 0.0821
## CLAY 0.790 -2.420 1.357 1.2698
## BERNARD -1.329 -1.612 -0.196 -1.9209
## YURKOV -0.869 0.433 -2.474 0.6972
## ZSIVOCZKY -0.106 2.023 1.305 -0.0993
## McMULLEN 0.119 0.992 0.844 1.3122# Cos2 of individuals
#:::::::::::::::::::::::::::::::::
# 1. square of the distance between an individual and the
# PCA center of gravity
center <- res.pca$center
scale<- res.pca$scale
getdistance <- function(ind_row, center, scale){
return(sum(((ind_row-center)/scale)^2))
}
d2 <- apply(decathlon2.active,1,getdistance, center, scale)
# 2. Compute the cos2. The sum of each row is 1
cos2 <- function(ind.coord, d2){return(ind.coord^2/d2)}
ind.cos2 <- apply(ind.coord, 2, cos2, d2)
head(ind.cos2[, 1:4])## PC1 PC2 PC3 PC4
## SEBRLE 0.00753 0.4975 0.08133 0.00139
## CLAY 0.04870 0.4570 0.14363 0.12579
## BERNARD 0.19720 0.2900 0.00429 0.41182
## YURKOV 0.09611 0.0238 0.77823 0.06181
## ZSIVOCZKY 0.00157 0.5764 0.23975 0.00139
## McMULLEN 0.00218 0.1522 0.11014 0.26649# Contributions of individuals
#:::::::::::::::::::::::::::::::
contrib <- function(ind.coord, comp.sdev, n.ind){
100*(1/n.ind)*ind.coord^2/comp.sdev^2
}
ind.contrib <- t(apply(ind.coord, 1, contrib,
res.pca$sdev, nrow(ind.coord)))
head(ind.contrib[, 1:4])## PC1 PC2 PC3 PC4
## SEBRLE 0.0385 5.712 1.385 0.0357
## CLAY 0.6581 13.854 6.460 8.5557
## BERNARD 1.8627 6.144 0.135 19.5783
## YURKOV 0.7969 0.443 21.476 2.5794
## ZSIVOCZKY 0.0118 9.682 5.975 0.0523
## McMULLEN 0.0148 2.325 2.497 9.1353

